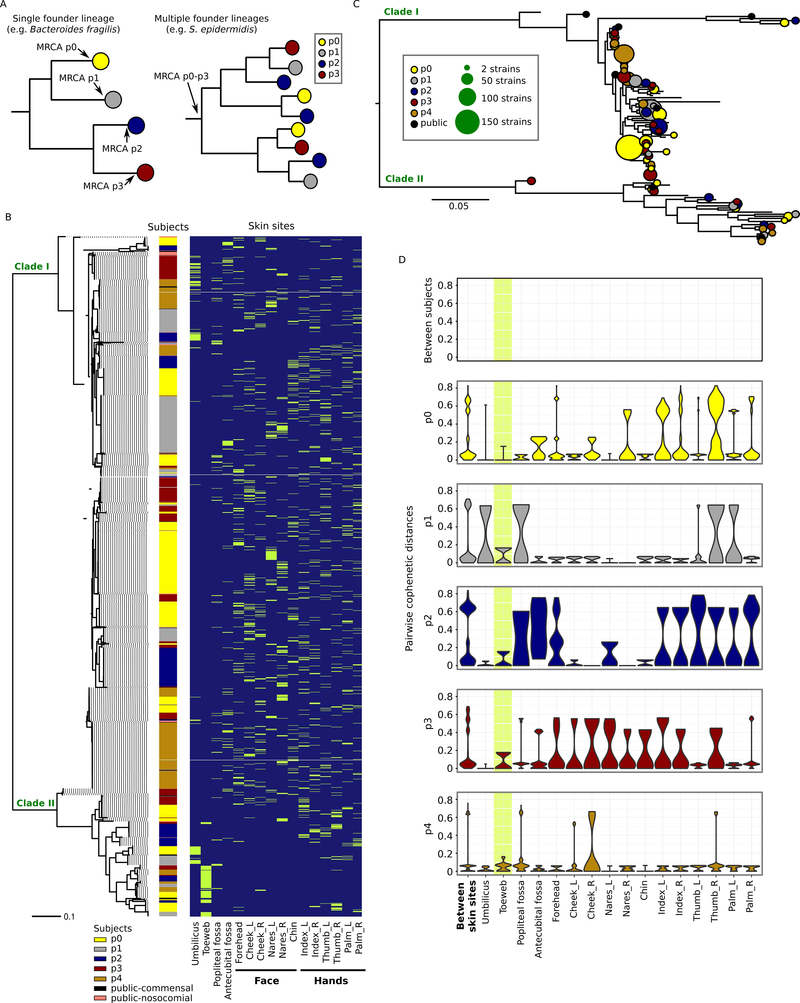
Figure 1.
Phylogenetic variation of the individualized S. epidermidis isolates. A, two alternative scenarios of within-individual evolution. Each circle represents a cluster of isolates diverged from a single founder lineage colonizing a given host. In the first scenario (left), all isolates from a given host diverged from a single founder lineage; in the second scenario (right), isolates from each host diverged from multiple distinct founder lineages. B, core-genome phylogeny (midpoint rooted) based on 58498 core-genome SNP loci for the 1482 isolates sampled in this study and 50 previously sequenced isolates from multiple diseased and healthy individuals. Skin site of each isolate is indicated in green. C, individualized S. epidermidis isolates evolved from multiple founder lineages. Each founder lineage is represented by a circle and is defined as the highest node from which at least 95% of the derived isolates (i.e. tip nodes) were either found in the same subject or public strains. The size of the circle represents the number of isolates derived from that lineage. D, pairwise cophenetic distances of the 1482 isolates. Note that the distribution of ‘between-subject’ distances depends on the sample size per subject, with p0, who had the most isolates cultivated, having the largest contribution. The toeweb is highlighted to illustrate its unusual between-subject similarity.