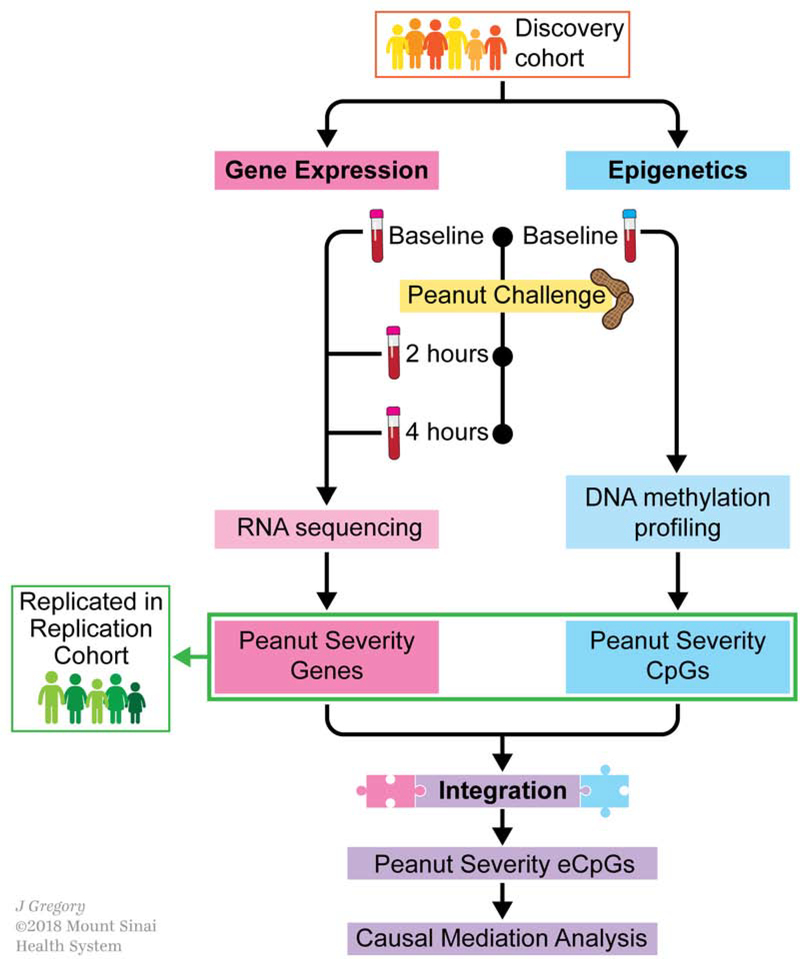
Fig. 1. Study design and analytical flow.
Schematic outlining the clinical sample collections, transcriptome and epigenome profiling, and main analyses. A discovery cohort consisting of 21 peanut allergic children underwent physician-supervised peanut challenge. For each subject, peripheral blood samples were drawn at baseline immediately before the peanut challenge, and at two hours and four hours after start of the peanut challenge, resulting in 3 samples per subject. To identify genes with expression changes over time associated with reaction severity (peanut severity genes), whole blood RNA sequencing for all samples and linear mixed-effects models were employed (pink box). To identify DNA methylation sites associated with reaction severity (peanut severity CpGs), CD4+ lymphocytes were isolated from each baseline peripheral blood sample, DNA methylation profiling was performed, and linear regression models were constructed (blue box). An independent cohort of an additional 19 peanut allergic children were then similarly profiled and used to replicate the peanut severity genes and peanut severity CpGs identified (green box). Integration of the transcriptome and epigenome results (purple box) was next pursued by building a combined network, identifying expression CpGs (eCpGs, i.e. CpGs associated with expression of peanut severity genes), and causal mediation analysis.