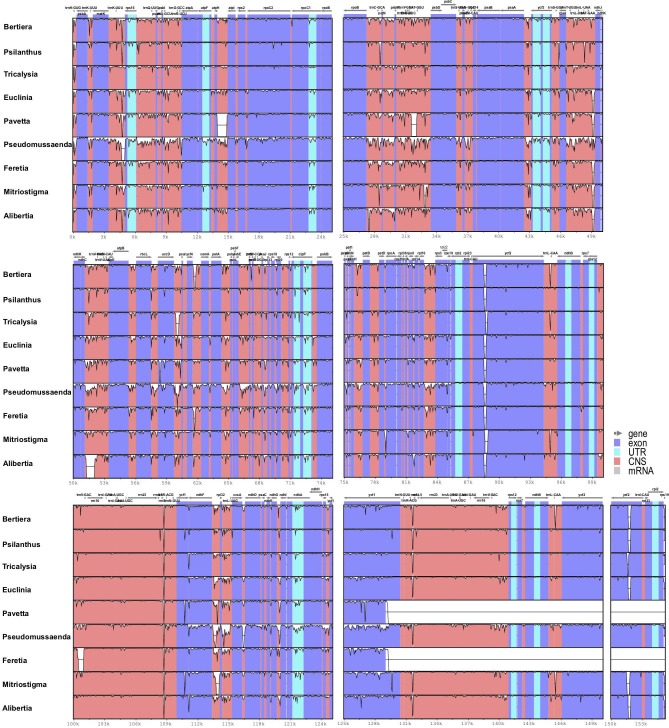
Fig 2. Sequence identity plot comparing nine species of subfamily Ixoroideae with Coffea arabica as the annotated reference genome using mVISTA.
The location and orientation of the genes are indicated on the top. Exons and UTRs are in purple and turquoise respectively. Conserved non-coding regions are in orange. The y-axis ranges from 100% (top) to 50% identity between each sequence and the reference. The order of the taxa used from top to bottom is: Bertiera iturensis (Bertiereae), Psilanthus ebracteolatus (Coffeeae), Empogona congesta (Coffeeae), Euclinia longiflora (Gardenieae), Pavetta schumanniana (Pavetteae), Pseudomussaenda stenocarpa (Mussaendeae), Feretia aeruginescens (Octotropideae), Mitriostigma axillare (Sherbournieae) and Alibertia edulis (Cordiereae). Feretia aeruginescens and Pavetta schumanniana showing only one IR in the current assembly.