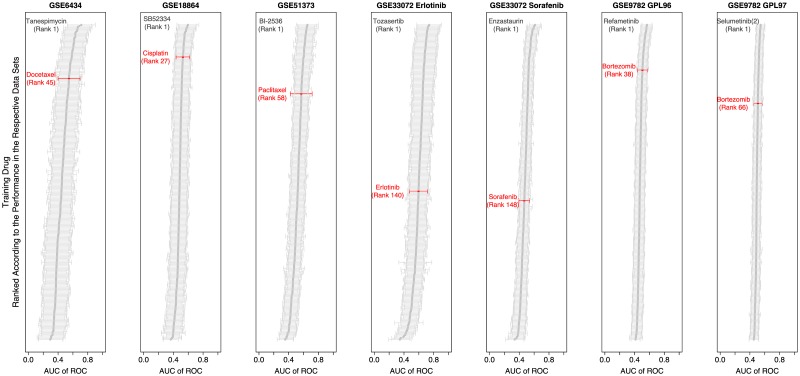
Fig 5. Drug specificity plot of FORESEE modeling pipelines.
Impact of the training drug on model performance for each of the seven different patient data sets: GSE6434, GSE18864, GSE51373, GSE33072 Erlotinib cohort, GSE33072 Sorafenib cohort, GSE9782 GLP96 cohort and GSE9782 GLP97 cohort. A set of 100 pipelines, which are listed in S1 Table, were randomly chosen and used to train translational models on the GDSC cell line data with each of the 266 drugs contained in the GDSC database individually and then tested on each of the patient data sets. For each of the data sets, the drugs are ordered with respect to the mean AUC of ROC of the 100 random pipelines trained with that drug. The red color marks the drug that is actually applied to the patient. The first-ranked drug is additionally indicated in order to facilitate the comparison of the different drugs and their modes of action. As an exception, for predicting GSE9782 GPL97 patient outcome, six pipelines that include RUV as homogenization method were not trained on those drugs that resulted in a training set that had more samples than features, as this was not compatible with the PCA step performed in this method.