Abstract
Two-component regulatory systems represent the major paradigm for signal transduction in prokaryotes. The simplest systems are composed of a sensor kinase and a response regulator. The sensor is often a membrane protein that senses a change in environmental conditions and is autophosphorylated by ATP on a histidine residue. The phosphoryl group is transferred onto an aspartate of the response regulator, which activates the regulator and alters its output, usually resulting in a change in gene expression. In this review, we present a historical view of the archetype EnvZ/OmpR two-component signaling system, and then we provide a new view of signaling based on our recent experiments. EnvZ responds to cytoplasmic signals that arise from changes in the extracellular milieu, and OmpR acts canonically (requiring phosphorylation) to regulate the porin genes and noncanonically (without phosphorylation) to activate the acid stress response. Herein, we describe how insights gleaned from stimulus recognition and response in EnvZ are relevant to nearly all sensor kinases and response regulators.
HISTORICAL ASPECTS OF EnvZ/OmpR SIGNALING
Signal transduction in bacteria is largely achieved by two-component regulatory systems. The first component is a sensor kinase (HK), usually a membrane protein, which senses an environmental signal and is phosphorylated by ATP on a conserved histidine residue. The second component is a response regulator (RR), which catalyzes the transfer of the phosphoryl group from the histidine of the HK onto a conserved aspartic acid residue. Most RRs are two-domain proteins, and phosphorylation of the N-terminal receiver domain alters the output of the C-terminal effector domain, which usually involves a stimulation of DNA binding. In some systems, the HK alters the level of the phosphorylated RR (RR∼P) by stimulating its dephosphorylation (referred to as phosphatase activity).
The HK EnvZ and its cognate RR OmpR are the products of the ompB operon and were historically identified as regulating the expression of outer membrane proteins OmpF and OmpC in response to changes in the osmolality of the growth medium (4, 5) (see Fig. 1). The operon is organized with ompR upstream, and its stop signal overlaps the start of envZ (6), with the result that there is substantially greater OmpR protein expressed relative to EnvZ (7). Approximately 50% of the two-component systems in Escherichia coli are organized similarly. Porin levels are influenced by a wide variety of growth conditions, including pH (8–11), temperature (12), osmolality (13), and growth phase (14–16). In general, the total amount of OmpF and OmpC remains relatively constant, but their levels are reciprocally regulated by environmental stimuli (13). For example, at low osmolality, OmpF is the major porin in the outer membrane, and at high osmolality, ompF transcription is repressed and OmpC becomes the major porin. The two porins differ from one another by their pore size and flow rates, with OmpC exhibiting a slower flux and a smaller pore (17). It is believed that sensing and adjusting the porins in the outer membrane is one strategy whereby E. coli and Salmonella recognize whether or not they are in a host environment (high osmolality) compared to a dilute environment (low osmolality).
Figure 1.
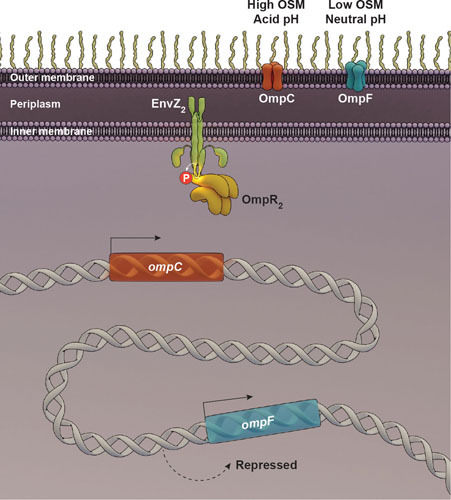
A classic view of EnvZ/OmpR regulation of outer membrane porins in response to osmotic or acid shifts. At low osmolality and neutral pH, OmpF is the major porin in the outer membrane. At high osmolality, ompF transcription is repressed and OmpC becomes the predominant porin. EnvZ autophosphorylation is stimulated by increasing osmolality, driving phosphotransfer to OmpR, dimerization, and high-affinity binding to DNA.
Domain Organization of EnvZ
EnvZ is composed of 450 amino acids and is located in the inner membrane. Two transmembrane domains (TM1 and TM2; residues 16 to 35 and 159 to 179) flank a 117-amino acid periplasmic domain (residues 36 to 158) (18). The cytoplasmic domain is divided into two functionally distinct subdomains (19, 20): a four-helix dimeric catalytic domain (amino acids 223 to 289), which contains the histidine that is phosphorylated (His243), and the OmpR binding region (inclusive of amino acids 269 to 276) (1, 3, 21). The ATP binding subdomain consists of amino acids 290 to 450. The catalytic domain is separated from TM2 by a HAMP (histidine kinase, adenylyl cyclase, methyl-accepting chemotaxis protein, and phosphatase) linker of approximately 43 amino acids. The HAMP linker is structurally conserved among many sensor proteins and may play a role in signaling in some HKs (22).
The catalytic domain forms a stable homodimer in solution with an apparent molecular weight of 19 kDa (23). A nuclear magnetic resonance (NMR) structure of the four-helix subdomain (20) provided the first structural insights of EnvZ at low osmolality (Fig. 2). The homodimer consists of a four-helix bundle with 2-fold symmetry. Each monomer contains two alpha helices, α1 (amino acids 235 to 255) and α2 (amino acids 265 to 286), that are separated by a 9-amino acid loop. Within the dimer, the helices are packed such that α1 is surrounded by an antiparallel α2. The core is hydrophobic, and the structure is conserved among HKs. The phosphorylated histidine, His243 lies in α1 and protrudes away from the helical bundle, where it is accessible to ATP from the ATP-binding subdomain. Within α1 is a region of dynamic instability (20) that is exquisitely sensitive to intracellular signals including acid pH (2, 3, 24) and high osmolality (1, 2). A soluble cytoplasmic construct of EnvZ (EnvZc; residues 180 to 450) lacking TMs 1 and 2, as well as the periplasmic domain, is fully capable of osmosensing and autophosphorylation in vitro and in vivo (1). More recently, others have identified additional HKs that are capable of intracellular signaling, although most of these systems are still lacking in mechanistic detail (25–27).
Figure 2.
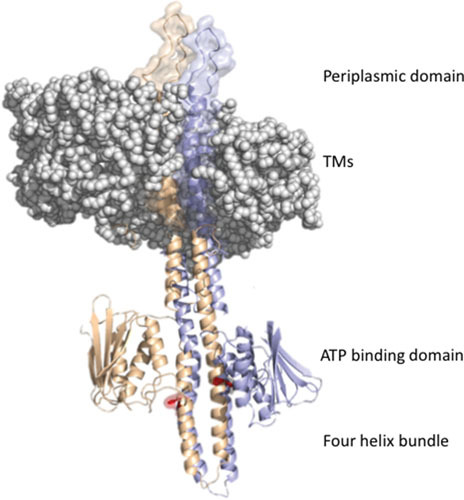
EnvZ domain structure. The periplasmic domain of an EnvZ dimer protrudes above the membrane, which is shown as a space-filling model (membrane from PDB ID: 3J00, EnvZ dimer frrom PDB: 4CTI) (190). The transmembrane domains (TMs) connect to the four-helix bundle formed from a dimer of two monomers (in purple and orange); a single His243 sidechain (phosphorylation site) is highlighted in red. The ATP binding domains flank His243.
The ATP binding domain is structurally similar to the ATP binding proteins DNA gyrase B and Hsp90. It is composed of an αβ sandwich composed of a five-stranded β-sheet (strand B: amino acids 319 to 323, D: 356 to 362, E: 367 to 373, F: 420 to 423, and G: 431 to 436) and three α-helices (α1: amino acids 301 to 311, α2: 334 to 343, and α3: 410 to 414). Between these two folds is a hydrophobic core that is highly conserved in HKs. Between α3 and α4, there is a “central loop,” which has no defined structure (Fig. 2). Binding of ATP occurs at α3 and the central loop and involves contacts with F and G β-strands. The central loop is near the ATP binding pocket formed by Asp-347, Asp-373, Ile-378, and Phe-387. The ATP triphosphate chain is exposed on the surface to allow transfer to His243 in the catalytic domain. Conserved glycines form G1 and G2 boxes located in the catalytic core and are essential for kinase activity.
Cis versus Trans Phosphorylation
Studies of the hybrid HK Taz, in which the extracellular aspartate receptor domain was fused with the EnvZ cytoplasmic domain (see “The Role of EnvZ Transmembrane and Periplasmic Regions” below) explored whether EnvZ phosphorylation was intra- (cis) or intermolecular (trans). Autophosphorylation of Taz was stimulated by aspartate binding (28). Dimerization promotes formation of a four-helix bundle, putting the sites of phosphorylation in each monomer in equal proximity to bound ATP from either monomer (20), making it equally plausible for cis or trans autophosphorylation in HKs. Single mutants that either lacked phosphorylation (H243V) or were impaired for ATP binding (a deletion of 146 amino acids of the C-terminal domain) were unable to activate ompC, but a mixture of the two restored signaling (29), suggesting that EnvZ phosphorylation occurs in trans.
While trans phosphorylation was proposed for EnvZ, biochemical studies of the Thermotoga maritima HK HK853 reported cis phosphorylation (30). The ability for HKs to undergo both cis and trans autophosphorylation was corroborated in the crystallographic structure of an EnvZ chimera containing the four-helix bundle subdomain of HK853 fused to the ATP binding subdomain of EnvZ (31). The ability to undergo cis or trans phosphorylation was dependent upon linker length, as well as the handedness of the helix. Substituting the EnvZ linker between α1 and α2 with the linker from PhoR switched phosphorylation from trans to cis phosphorylation (30, 31). Thus, both cis and trans phosphorylation pathways are structurally and biochemically possible in EnvZ and its homologues.
Regardless of the mode of autophosphorylation, two functional groups impact His243 autophosphorylation (see “A Combination of Side Chain and Peptide Backbone Effects Drive Signaling” and “What Activity is Regulated by Osmolality/Acid pH?” below). Asp244 functions as a base to abstract a proton from Nδ and promotes nucleophilic attack on the γ-phosphate of ATP (31). Crystallographic analysis offers a snapshot of a fully ON (high osmolality) or OFF state (low osmolality), but amide hydrogen deuterium exchange mass spectrometry (HDXMS) provides direct insights into the ensemble behavior of EnvZ in solution, which is critical for function (1). In low osmolality, His243 is positioned by a coordinated relay of interactions from the backbone carbonyl of Ala239 (one helical turn above His243) through the imidazole side chain of His243 to the carboxylic acid side chain of Asp244. The interplay of these two flanking moieties regulates His243 tautomerization, rotamerization, and phosphorylation. The intrinsic disorder within the locus containing the His243 side chain affords a low basal level of autophosphorylation (32). High osmolality stabilizes the helical backbone and relieves the Ala239 carbonyl link to enable anchoring of the His243 imidazole by a H-bond with Asp244 at the Nδ of His243, enabling phosphotransfer to the Nε of His243. Thus, the core of osmosensing is a His-Asp/Glu dyad positioned within a flexible helix.
The Role of EnvZ Transmembrane and Periplasmic Regions
It was assumed that because EnvZ was an inner membrane protein, the TM and periplasmic domains were important for its function. HAMP domains have been shown to be important for relaying the effects of extracellular stimuli (22, 33, 34). The periplasmic domain shows the greatest diversity in length and sequence among EnvZ homologues from different bacterial species. These range from 6 residues in Xenorhabdus to 147 residues in E. coli. Chimeras of E. coli EnvZ with the periplasmic domain from Xenorhabdus restored full functionality, offering evidence that the cytoplasmic domain was pivotal for osmosensing and that the TMs were not essential (35). The integrative capacity of the cytoplasmic domain when coupled to alternative signaling domains was further inferred from functional chimeras of the N-terminal segments of the aspartate receptor (Tar) fused with the cytoplasmic domain of EnvZ (Taz1) (28). Because signaling occurred when the EnvZ TMs or periplasmic domain was absent or replaced to construct a chimera, these approaches ruled out an essential role for the EnvZ periplasmic domain and the TMs in EnvZ signaling. An N-terminal truncation of EnvZ (EnvZ115) that lacked TM1 and did not localize to the inner membrane did not respond to osmolality shifts. It was therefore deduced that EnvZ required its transmembrane domains for signaling (36). However, EnvZ115 ended up in inclusion bodies (37), and thus it was unable to respond to extracellular or intracellular signals. In contrast, the soluble, cytoplasmic construct EnvZc was fully capable of osmosensing by responding to signals that arose in the cytoplasm (1).
How Do Extracellular Signals Translate into Intracellular Signals?
In response to an increase in extracellular osmolality, a passive diffusion of water out of the cell occurs, and cells reduce their volume as the intracellular water activity drops (38). This reduction in cell volume is evident in super-resolution images of E. coli at high osmolality (39). If it is available, extracellular K+ ions are taken up via the Trk channels (40), which are stimulated by the Kdp two-component system (41). Synthesis of organic osmolytes such as glycine betaine is stimulated and subsequently replaces the high levels of intracellular potassium (42). Measurements of intracellular osmolytes identified the large changes in osmolality that can be endured by Gram-negative bacteria such as E. coli and Salmonella (up to 1,800 mOsm Kg–1) (43). These results highlight the unique properties of bacteria compared to eukaryotes, which tightly regulate both osmolality and pH to be isosmotic with blood (290 mOsm Kg–1, pH 7.35 to 7.45). In response to extracellular osmotic stress, the bacterial cytoplasm becomes concentrated (43), and secondarily, a decrease in intracellular pH occurs (2, 3).
Freely Diffusible Osmolytes Do Not Stimulate EnvZ
Diffusing osmolytes such as glycerol do not stimulate EnvZ, and now we understand the basis for this finding. Osmolytes are small organic compounds that exert a dramatic influence on the protein folding reaction without making or breaking covalent bonds. Protective osmolytes push the folding equilibrium toward N, the native state, by raising the free energy of the unfolded state (U), favoring the folded population. Denaturing osmolytes push the equilibrium toward U, the unfolded state. The osmolyte effect operates predominantly on the peptide backbone, a component common to all amino acid residues in a protein (44, 45). The backbone transfer of free energy (Δgtr) primarily determines the extent to which osmolytes stabilize (Δgtr > 0) or destabilize (Δgtr < 0) the protein relative to an equivalent aqueous solution. Interactions between the peptide backbone and osmolytes are dominated by the surface area and outer-group polarity of the osmolyte (46). Sucrose has a larger polar surface area compared to glycerol and a larger Δgtr (56 cal/mol versus 22 cal/mol). Glycerol is ranked as the weakest of protecting osmolytes on the transfer energy scale (46). Thus, osmolytes such as sucrose and betaine, for example, exert much greater effects on peptide backbone stabilization of EnvZ, which then activates autophosphorylation (see “What Activity is Regulated by Osmolality/Acid pH?” below).
Lipid Allostery Couples EnvZ Signaling and Activation, the Effect of Procaine, Robustness
The observation that EnvZc was capable of osmosensing (1) raised the obvious question as to whether the TMs or the periplasmic domain or the lipid composition of the membrane contributed to EnvZ function. Earlier studies established that the four-helix bundle subdomain was sensitive to osmolytes and insensitive to nucleotide binding. In contrast, the ATP binding subdomain was sensitive to nucleotide binding but insensitive to osmolytes (1). Membrane lipids serve to couple these two domains. A notable change in HDXMS of the full-length solubilized EnvZ incorporated into nanodiscs compared to EnvZc was in the region of the Gly-rich motif in the ATP binding site (32). In the presence of lipids, the rate of ATP hydrolysis in the nucleotide binding domain increased, which was evident as higher turnover. Thus, while EnvZc forms the core of the osmosensing locus, membrane anchoring of EnvZ enables an interaction of the ATP binding domain with the membrane that enhances autophosphorylation at His243. Thus, ATP hydrolysis is coupled to osmosensing. This effect of lipids likely forms the basis for how EnvZ/OmpR responds to procaine (47) and other membrane perturbants (48). These agents alter the lipids, which then affect interactions with the glycine-rich loop of the ATP binding domain, stimulating ATP turnover and subsequent phosphorylation at His243 (32). Stimulation of EnvZ autophosphorylation would lead to repression of ompF and activation of ompC (47–49). Regions of full-length EnvZ that showed increased deuterium exchange included the HAMP, kinase, TM, and periplasmic domains (32). Inside bacterial cells, overexpression of EnvZc resulted in a higher level of β-galactosidase activity of an ompC-lacZ fusion compared to the wild-type protein (1). This was not surprising, since HKs are typically expressed at very low levels, and overexpression enhances activity (1). This finding suggests that porin regulation by EnvZ/OmpR is not robust. An early proposal that OmpR∼P levels were independent of the EnvZ concentration argued that the EnvZ/OmpR system was robust (50). Our findings conflict with claims of robustness because in one scenario, ompC-lacZ activity increased at elevated EnvZc concentrations (1). In a separate experiment, higher levels of EnvZ overexpression inhibited ompC transcription, and the number of EnvZ molecules produced was verified by molecular counting using super-resolution imaging (21).
EnvZ Biochemical Activities Underlie Signaling
The kinase activity results from EnvZ binding ATP and its subsequent autophosphorylation of His243 (51). At high osmolality, autophosphorylation of His243 is enhanced (1). EnvZ then transfers the phosphoryl group to OmpR at its conserved aspartic acid (Asp55) (52). This phosphorelay is at the heart of two-component signaling (Fig. 3). EnvZ signals through OmpR, controlling the concentration of OmpR∼P to control porin gene expression (5). The first step in the signaling cascade thus involves osmolyte stimulus recognition by EnvZ, leading to enhanced autophosphorylation of His243. At high EnvZ concentrations in vitro, EnvZ can stimulate the dephosphorylation of OmpR∼P (referred to as a phosphatase activity). Thus, EnvZ can modulate the level of OmpR∼P by altering any or all of these reactions.
Figure 3.
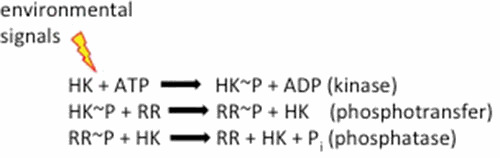
Reaction scheme of EnvZ-OmpR two-component signaling. EnvZ binds ATP and is autophosphorylated at His243. Phosphorylated EnvZ transfers the phosphoryl group to OmpR. OmpR∼P binds with higher affinity to the porin promoters and activates transcription. At high concentrations, EnvZ can catalyze OmpR∼P dephosphorylation.
EnvZ Mutants with Altered Biochemical Activities
Mutations have been isolated in envZ that alter its phosphorylation, phosphotransfer, and dephosphorylation activities, although complete uncoupling of these activities has not effectively been demonstrated, apart from the requirement of His243 for autophosphorylation (51). The interpretation of the various results highlights the differences in approach and the “conflict” between geneticists and biochemists (53, 54). Biochemists complain that genetic experiments are indirect, while geneticists argue that in vitro experiments do not necessarily recapitulate what happens in vivo. Localized mutagenesis of envZ identified two substitutions, one that was one helical turn away from the phosphorylated histidine, A239T, and a periplasmic substitution, P159S. These substitutions were described as kinase minus, phosphatase plus (K–P+) (55, 56), but direct measurements revealed that the A239T substitution exhibited a 3-fold lower affinity for ATP and was also impaired in phosphotransfer to OmpR, indicating multiple effects on EnvZ activities from the single substitution (37).
EnvZ Dephosphorylation of OmpR∼P
In many two-component systems, dephosphorylation of the RR∼P via the HK (the so-called phosphatase activity) limits the level of the activated RR and resets the system. In some systems, it is clear that RR∼P dephosphorylation occurs or is stimulated by accessory proteins. For example, PII-stimulated dephosphorylation of NtrC∼P (57, 58) and CheY∼P dephosphorylation by CheZ (60, 61). In the case of EnvZ, it is poised to increase autophosphorylation in response to signaling (i.e., OmpR∼P levels are relatively low in the absence of envZ) (5), whereas its close homologue CpxA is poised to regulate CpxR∼P turnover (i.e., CpxR∼P basal levels are high in the absence of cpxA) (59, 62). Kinetic studies proposed that the HK phosphatase activity limits cross-talk between highly homologous two-component systems (63), but more recent studies of covarying residues (64) and our HDXMS experiments determined that what limits cross-talk is the sequence specificity of the RR binding domain of the HK (1, 32). It is important to note that concentrations of OmpR far exceed those of EnvZ in E. coli and RRs in general compared to HKs (7). Therefore, all of the EnvZ would be complexed with OmpR. The significance of this complex is that osmolyte stimulation of EnvZ phosphorylation and phosphotransfer from EnvZ to OmpR becomes a unidirectional process in vivo (see Fig. 4). Experiments performed with kinase null strains alter the free concentration of the RR in the cell because it is no longer complexed with the HK. The observed OmpR∼P “phosphatase” activity of EnvZ is in part a limitation of in vitro biochemical experiments performed with a large excess of EnvZ (63, 65). When the assay was performed with EnvZ at concentrations similar to the in vivo ratios of EnvZ to OmpR, turnover was very slow (66) (see reference 67 for a review). While we do not discount the ability of EnvZ to dephosphorylate OmpR∼P when it is in substantial excess, the relative concentrations of EnvZ and OmpR in vivo render the phosphorylation of OmpR unidirectional from EnvZ to OmpR in vivo. A similar behavior is likely in many, but not all, two-component systems. An important distinction may be those systems in which regulation is linked to central metabolism, as is the case with CpxA/R (62).
Figure 4.
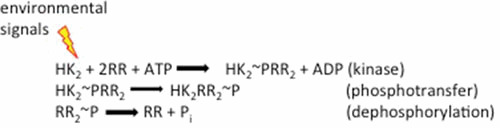
A modified reaction scheme of two-component signaling. Because the RR is in great molar excess compared to the HK, all of the HK would be complexed in vivo. ATP binds to the HK and autophosphorylates, driving phosphoryl transfer and dimerization of the complexed RR. The RR∼P is then stimulated to activate its downstream pathway, usually through enhanced DNA binding. The phosphorylated RR dimer can undergo dephosphorylation to then rebind to the HK.
A demonstrated role for the phosphatase activity in signaling is based on events that occur in the absence of the HK, which as mentioned above, alters the RR free concentration in the cell. Acetyl phosphate (AcP) can phosphorylate RRs in vitro and in vivo (62, 68–70), although RRs vary in their ability to be phosphorylated by AcP. CpxR is an RR that is more readily phosphorylated by AcP than OmpR (our unpublished observations; 72, 73). These differences are evident in the low activity of an ompF-lacZ reporter in an envZ null strain (55). In contrast, a cpxA null strain showed much higher activity of CpxR regulation of degP- and cpxP-lacZ transcriptional fusions (74), suggesting a higher concentration of CpxR∼P produced by AcP. A gain of function mutant (Cpx*) (75, 76) located approximately one helical turn below the phosphorylated histidine was described as a K+P– mutant, but it clearly has severely reduced autophosphorylation and phosphotransfer activities as well (74). Growing cells in conditions that alter AcP levels (such as acetate or pyruvate) would also influence the cellular concentration of phosphorylated RRs (71). It is also worth noting that many of these measurements were made on protein fusions to maltose binding protein, which can dramatically affect protein:protein interactions, as well as rates of phosphorylation and phosphotransfer. In particular, we have observed substantially lower levels of OmpR∼P produced by maltose binding protein-EnvZ compared with other EnvZ constructs or from AcP phosphorylation. One interpretation of experimental observations is that the HK phosphatase activity is important for suppressing cross-talk with other two-component systems. An alternative interpretation is that the presence of the HK complexes the RR and reduces its availability as a substrate for phosphorylation by AcP. Thus, when the cognate HK is missing, the free RR concentration increases, and it can be a substrate for AcP (Fig. 4). Single-molecule localization microscopy experiments may help to identify such HK/RR complexes and determine whether they are simultaneously complexed with DNA in vivo and whether complex formation is influenced by environmental stress.
Strain differences are also likely to play a role in the observed behavior of EnvZ in response to environmental stresses, which also affects the interpretation of mutant behaviors. The majority of experiments on the EnvZ/OmpR system were conducted on the E. coli strain MC4100, which responds differently to extracellular acid or osmotic stress than the probiotic Nissle strain (77, 78), and on MG1655, a sequenced strain (79). It appears that the large deletions that occurred during the construction of MC4100 for making lacZ transcriptional fusions may make it difficult to generalize results of physiological studies to other strains (80). For example, in response to acid or osmotic stress, MG1655 acidifies from an initial intracellular pH of 7.13 to 6.55 or 6.75, respectively. MC4100 was slightly acidified initially compared to MG1655 (pH 6.80 versus pH 7.13) (3), but MC4100 did not acidify further in response to acid stress (3). Instead, MC4100 maintained its cytoplasmic pH throughout the experiment (internal pH = 6.83). This was the case whether the external pH was 7.2, 5.6, or induced by the osmolyte sucrose. The addition of osmolytes increased the pHi of MC4100 from 6.8 to 7.15, as reported (81). This was opposite to what we observed with E. coli MG1655 (3). Differences in intracellular pH resulting from environmental stress are likely to have significant effects on gene regulation, since histidyl phosphates are rapidly hydrolyzed under acidic conditions (3, 82), and acid pH alters the conformation of the HK (see “A Combination of Side Chain and Peptide Backbone Effects Drive Signaling” and “What Activity is Regulated by Osmolality/Acid pH?” below).
WHAT ACTIVITY IS REGULATED BY OSMOLALITY/ACID pH?
A long-standing question in the field was which activity of EnvZ was regulated by osmotic stress? A study by Jin and Inouye proposed that at high osmolality, OmpR∼P levels increased as a result of a decrease in the phosphatase activity (83). Their hypothesis was based on experiments with the chimeric kinase Taz (28), which contains the periplasmic domain of the Tar aspartate chemoreceptor fused to the cytoplasmic domain of EnvZ. Taz activates ompC in the presence of aspartate. Although activating EnvZ using a known ligand was clever, the construct has several limitations that cast doubt on whether conclusions based on Taz are physiologically meaningful. The problem is that the domains derived from Tar behave differently from native Tar, so how does that impact the interpretation of the function of the EnvZ domains? For example, Taz requires 1 to 5 mM aspartate to activate ompC compared to Tar, which binds aspartate with a Kd of 1.2 μM (84). Maltose also activates Tar, but it did not enhance ompC transcription, and finally, aspartate did not affect ompF transcription. More recent studies have shown that EnvZ phosphorylation is directly stimulated by osmolytes, including K+ (85), Na+, and sucrose (1). Furthermore, HDXMS also demonstrated that EnvZc could sense osmolality without being in the membrane and that the source of the signal was cytoplasmic (1) (see “The Effect of Osmolytes or Acid pH on EnvZ” below). Thus, it is now established that the major effect of osmolality is to stimulate EnvZ kinase activity (see also “Activation of OmpR” below).
MzrA
MzrA was identified by a genetics approach as a modulator of the EnvZ/OmpR system (86). MzrA is a 127-amino acid periplasmic protein that interacts with EnvZ to modulate its kinase activity and elevate OmpR∼P levels. Subsequent experiments determined that the periplasmic domain of MzrA, and not its single transmembrane domain, was the site of interaction with EnvZ (87). A construct that lacked the TM retained activity, indicating that TM localization was not essential. The role of MzrA appears to be important in envelope stress rather than osmotic stress. During envelope stress, the Cpx and σE pathways are activated, MzrA concentrations increase, and ompF is repressed via the action of OmpR∼P (86, 87). Reducing OmpF levels appears to alleviate the ensuing envelope stress.
Cross-Talk Between Noncognate HK-RR Pairs
Cross-talk between noncognate HK-RR pairs is suppressed by a high degree of sequence specificity encoded in the RR binding site on the HK (64). In EnvZ, this region encompasses residues 267AESINKDIEECN278 (1). Labeling of Cys-277 with a fluorophore prevented OmpR binding, supporting the AESINKDIEECN locus as a site of EnvZ/OmpR interaction (21). The closest homologue to OmpR is CpxR. In the absence of envZ and cpxR, CpxA can phosphorylate OmpR (72), increasing OmpR binding to ompF and ompC sites (88). This activity is entirely suppressed in a wild-type EnvZ strain. Why? We hypothesize that the presence of the cognate HK has higher affinity/specificity for its cognate RR, preventing a lower-affinity interaction with CpxR. Now that there are new tools and super-resolution imaging in bacteria is possible (39, 89, 90), it will be worthwhile to determine the extent to which OmpR is bound to EnvZ using single particle tracking photoactivation localization microscopy (spt-PALM) and compare with OmpR binding to CpxA in an envZ null strain. Presumably, an OmpR interaction with its cognate kinase EnvZ is favored, and we would expect to see a higher population of bound OmpR molecules in the EnvZ-labeled strain than in the envZ null, CpxA-labeled strain.
Ensemble View of EnvZ Signaling
HDXMS has provided an overview of conformational dynamics of EnvZ in solution, where the four-helix bundle subdomain serves as a primary signaling module for changes in osmolality and pH (1). His243 is positioned within a highly disordered segment of a longer helix and flanked by a helix that is sensitive to OmpR binding (see “‘An Asp for an Asp’ Drives Phosphotransfer” below). Together, these disordered helices adopt an ensemble of multiple conformations of EnvZ in solution. Osmolytes and environmental stimuli function to alter this ensemble to trigger nondiscrete signaling outputs. The four-helix bundle is a point of convergence of multiple signaling inputs, including signals from membrane lipids (see “Lipid Allostery Couples EnvZ Signaling and Activation, the Effect of Procaine, Robustness” above) and specific OmpR interactions (see “‘An Asp for an Asp’ Drives Phosphotransfer” below). This provides EnvZ with a wide dynamic range for responding with high sensitivity to gradients of osmolytes as well as pH stimuli. Convergence of multiple signals from the TMs and the EnvZ periplasmic domain (including MzrA; see “MzrA” above), as well as the membrane, explains the allostery observed with the Tar-EnvZ chimera Taz (28), the activating effects of a periplasmic mutant EnvZ P151A (56), and the lipid effects on the ATP binding domain (91).
THE OmpR SUBFAMILY OF RESPONSE REGULATORS
OmpR is a two-domain RR with an N-terminal receiver (phosphorylation domain) and a C-terminal DNA binding domain (92). The C-terminal effector domain is a winged helix-turn-helix motif that binds DNA (93–95). OmpR is an extensively studied member of a subfamily of RRs that includes PhoB, VirG, ResD, and CpxR, among others. RRs are divided into subfamilies based on the structure of their DNA binding domains. The OmpR subfamily has 14 homologues in E. coli alone (96). The receiver domain is highly conserved among RRs and consists of five alpha helices surrounded by a central β-sheet of five parallel β strands (97; Fig. 5A). Several critical invariant amino acids are in the loops between α helices and β strands: between β4 and α4 is a threonine (Thr83 in OmpR) (98), between β5 and α5 is a tyrosine (Tyr102), and the aspartic acid that is phosphorylated is in the loop between β3 and α3 (Asp55). Phosphorylation occurs at only this one site (68), Asp55 (52).
Figure 5.

(A) A highly conserved receiver domain (phosphorylation site) structure. Key residues that contribute to phosphorylation are highlighted. (B) Ribbon diagram of the NMR structure of the C-terminal domain of OmpR in one orientation (left) and rotated 90° (right) (reprinted with permission from reference 95).
OmpR binds as a dimer to multiple DNA sites at the ompF and ompC promoters between –100 and –60 upstream of the transcription start site (98, 99). Repression of ompF involves an additional upstream binding site between –384 and –354 (100). The crystal structure of the C terminus of OmpR (OmpRc) was determined independently by two groups (94, 101). The structures revealed the presence of a winged helix-turn-helix motif (wHTH) in the DNA binding domain of OmpR (see Fig. 5B). The recognition helix is unusually long and is the major determinant of DNA binding specificity. The turn of the HTH is also unusually long. Most canonical HTH proteins have turns of 3 to 4 amino acids, whereas OmpR subfamily members have turns of 10 amino acids (102). The turn is dynamic, and substitutions in the turn affect interactions with RNA polymerase (RNAP) (103–105). In the case of PhoB, there is strong evidence that the turn of the HTH interacts with the σ subunit of RNAP, but in the case of OmpR, substitutions in the turn affect DNA binding (D. Walthers and L. J. Kenney, unpublished results).
OmpR is somewhat unusual among response regulators in that the unphosphorylated protein has quite high affinity for DNA (Kd of ∼150 nM) (106), while the Kd of the phosphoprotein is 5 nM. These measurements were made possible by our ability to separate phosphorylated and unphosphorylated forms of the protein by high-pressure liquid chromatography (106), an approach that is now widely used with other RRs. High-affinity binding by OmpR/OmpR∼P is also observed in in vitro transcription studies (89, 107), as well as by atomic force microscopy (3) and super-resolution imaging (91). OmpR makes more phosphate backbone contacts and fewer specific base contacts (108) than RRs that recognize precise motifs such as PhoP, PhoB, and others. This lack of specificity enables OmpR to contribute to acid and osmotic stress responses (2, 3, 24, 109) and to evolved virulence gene regulation in Salmonella (110, 111) (see “Virulence” below). Until recently, it was believed that unphosphorylated OmpR did not play a role in gene expression (5), but it is now appreciated that OmpR can act noncanonically to regulate genes outside of the porin regulon that facilitate survival and growth in response to acid and osmotic stress (see “Noncanonical Activation of OmpR” below) (2, 3, 24).
One of the unique features of OmpR subfamily members is the presence of a four-stranded β-sheet at the N terminus of the effector domain (see Fig. 5B). A model whereby phosphorylation of the receiver domain results in subtle conformational changes that are transmitted to the β-sheet of the C terminus was proposed based on the cocrystal structure (112, 113). Cysteine-scanning mutagenesis through this region in OmpR suggested that the β-sheet may be involved in oligomeric interactions (114), and the role of the β-sheet may vary in function among OmpR subfamily members.
Activation of OmpR
Phosphorylation of EnvZ at His243 from ATP subsequently leads to the phosphorylation of OmpR at Asp55 (52). Small-molecule phosphodonors such as acetyl phosphate or phosphoramidate also phosphorylate OmpR at the same locus (68). Phosphorylation of OmpR in its N-terminal receiver domain increases the affinity of its C-terminal domain by at least 10-fold for the regulatory regions upstream of the ompF and ompC genes (106). The communication between OmpR domains is bidirectional; the presence of high-affinity ompF or ompC sites stimulates OmpR phosphorylation in vitro. These experiments led to the proposal that OmpR might be activated while bound to its target DNA. This would require the formation of a complex between the membrane-bound EnvZ with OmpR, while it is physically bound to DNA. Genetic evidence for such an HK-RR-DNA complex exists, both in the EnvZ/OmpR system (115) and in its close homologue CpxA/R (Champion and Silhavy, personal communication). In OmpR, two substitutions, G94D and E96A, eliminate ompF and ompC activation, and the G94D substitution is dominant (115, 116). These mutants are of interest because EnvZ was required for their dominance. At the time, the authors had no explanation for this surprising finding. Both mutants were unable to bind DNA, suggesting that oligomerization is an essential prerequisite to binding. These mutants lie on the face of OmpR that is opposite the phosphorylation site. Both were capable of phosphorylation, and it was proposed that the mutants were unable to oligomerize after phosphorylation. This interpretation absolutely fits with a recent observation that OmpR dimerization required EnvZ but did not require phosphorylation (see “Noncanonical Activation of OmpR” below) (3).
A four-state model has been proposed in which OmpR exists as an equilibrium mixture between four distinct states (Fig. 6). Free OmpR in the unphosphorylated form (Fig. 6A), OmpR∼P (Fig. 6B), the unphosphorylated form bound to DNA (Fig. 6C), and OmpR∼P bound to DNA (Fig. 6D). The reaction step that is most affected by DNA depends on the phosphodonor. In the presence of AcP, DNA binding stimulated OmpR phosphorylation dramatically (about 25-fold faster), with little or no effect on OmpR∼P dephosphorylation (117). In Fig. 6, this means that C to D is much faster than A to B. DNA binding also slowed dephosphorylation about 2-fold; i.e., D to C is slower than B to A. When phosphorylating with EnvZ, D to C was much slower than B to A. In either case, the overall effect of DNA was to increase the net rate of OmpR∼P formation by about 50-fold. Qin and coworkers proposed that OmpR∼P binding to DNA made it inaccessible to dephosphorylation by EnvZ (118). However, this does not fit with their proposal that the EnvZ phosphatase activity was the step that was regulated by high osmolality (83) (see “What Activity Is Regulated by Osmolality/Acid pH?” above). According to Qin et al., OmpR∼P bound to DNA would not be able to be dephosphorylated, raising the question of how discrimination between OmpR∼P bound to ompF (presumably at low osmolality) and OmpR∼P bound to ompC (at high osmolality) and differential regulation would occur.
Figure 6.
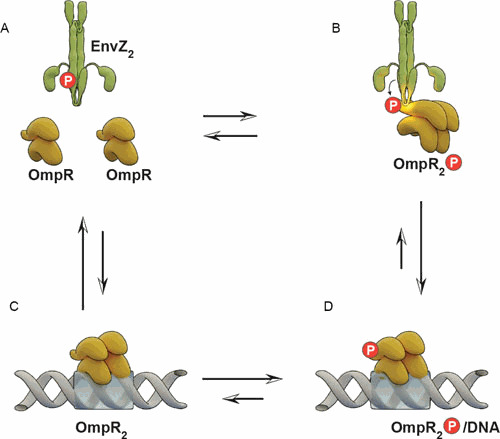
A reaction scheme of events that drive OmpR phosphorylation and DNA binding. OmpR is a two-domain protein; the N-terminal phosphorylation or receiver domain is joined to the C-terminal DNA binding effector domain via a flexible linker. (A) The protein is shown in the uncomplexed state (B) phosphorylated, (C) bound to DNA in the unphosphorylated state, and (D) bound to DNA while phosphorylated. The arrows depict transitions between these four states. Further complicating the scheme is the dimerization of OmpR (not depicted for simplicity). OmpR is a monomer in solution, and phosphorylation or interaction with EnvZ drives dimerization. In panels B, C, and D, the conformation of the linker is altered by phosphorylation, DNA binding, or both events.
These questions were addressed in studies using fluorescence anisotropy to measure EnvZ/OmpR interactions. OmpR was labeled at its N-terminus with fluorescein, and EnvZc was added to the mixture. The Kd for EnvZ binding to unphosphorylated OmpR was 425 nM, and binding was unaffected by the addition of ompF or ompC DNA (66), indicating that DNA binding did not affect the ability of OmpR to interact with EnvZ. Phosphorylation of OmpR reduced its affinity for EnvZ to unmeasurable levels, to at least 10-fold lower affinity (i.e., >5 μM). This finding is inconsistent with a proposed role for DNA in the OmpR/OmpR∼P equilibrium in which DNA prevents EnvZ from dephosphorylating OmpR∼P (118), because the affinity of EnvZ for OmpR∼P is already too low (66). The binding measurements are consistent with measurements in the chemotaxis system in which phosphorylation of the RR CheY (an OmpR homologue) reduced its affinity for the CheA kinase (119). The reduced affinity of CheY∼P for CheA would promote its binding to the switch proteins of the flagellar motor. Similarly, phosphorylation of OmpR by EnvZ∼P while it is bound to DNA (Fig. 6C) would reduce its affinity for EnvZ and promote an interaction between OmpR∼P and RNAP. Because the cellular concentration of EnvZ is so low, a Kd of >5 μM for EnvZ/OmpR∼P would mean that these two partners would hardly ever interact, suggesting that EnvZ cannot be the primary mechanism of OmpR∼P dephosphorylation. Thus, we favor a view where OmpR∼P turnover occurs rapidly enough in vivo, due to the intrinsic instability of aspartyl phosphate intermediates, eliminating the need for phosphatase activity in controlling signaling. Recent phosphorylation results clearly demonstrated the link between increasing osmolality and increasing levels of EnvZ∼P (1), suggesting that the kinase activity was the step regulated by environmental signals. Of course, an ability to actually measure the level of phosphoproteins in vivo would be enormously helpful in providing a quantitative molecular picture of signaling events.
OmpR Binding Sites
OmpR binds to three tandem sites immediately upstream of the –35 element (see Fig. 7) between approximately –100 and –40 from the transcriptional start site at both the ompF and ompC promoters (99, 120–122). At ompF, there is an additional upstream site, F4, that is required for repression (123). A genetic approach was employed to determine which OmpR binding site(s) participates in the recruitment of RNAP (124). DNA fragments containing the binding sites were used in a gel retardation assay with purified OmpR, EnvZ, and ATP. Only the DNA fragments containing the site between –100 and –80 of either promoter were shifted in the presence of protein and ATP (F1 and C1 sites; see Fig. 7). The interpretation was that F1 and C1 each comprise high-affinity OmpR binding sites. The F1 and C1 fragments were cloned in either orientation upstream of the ompC –35 element. Both fragments were able to direct transcriptional activation when present in either direction, but in both cases, the forward-facing F1 or C1 fragments were more active. Insertion of a 4-bp spacer between the binding site and the –35 element abolished transcriptional activation. Insertion of a 10-bp fragment at either promoter reduced activation, but the level of transcription was still considerably higher than the control strain (124). The interaction between OmpR and RNAP must be flexible, provided that the proper helical phasing of each protein bound to DNA is maintained. Thus, OmpR bound immediately upstream of the –35 element or at one helical turn upstream is sufficient to activate transcription, indicating that at the ompF or ompC promoter, occupancy of the F3 or C3 sites is required for activation (98). A prediction from these studies is that OmpR bound to DNA exists as a head-to-head or tail-to-tail dimer (114). This is intriguing, because the OmpR binding sites are asymmetric, and protein dimers in head-to-head or tail-to-tail binding modes typically bind to inverted repeat elements. A previous study reported a head-to-tail mode of binding based on Cu-phenanthroline cleavage of OmpR bound to DNA (125). These conflicting results likely will be reconciled by the cocrystal structure of OmpR bound to ompF/ompC DNA. Although OmpR exists as a monomer in solution, dimerization occurs upon phosphorylation (126) or after interaction with EnvZ (3). Sequence requirements for binding to the downstream half site appear to be less stringent than for initial binding to the upstream half site, implying that at least some OmpR binding sites may actually contain a poorly defined inverted repeat.
Figure 7.
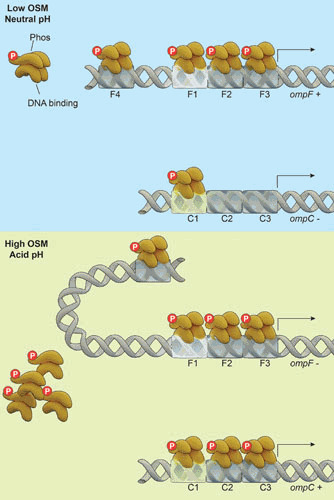
The porin promoters. OmpR∼P binds as a dimer to sites located between –100 and –40 upstream of the transcription start site. The sites between –100 and –80 (F1 and C1) are the highest affinity for OmpR and OmpR∼P. Phosphorylation is required for occupancy of the lower-affinity sites F2-F3 and C2-C3. An additional site at ompF (F4) is required for repression between –380 and –350, and repression is predicted to occur via loop formation assisted by the DNA bending protein IHF. Presumably, the loop then occludes and prevents RNA polymerase binding and subsequent activation of transcription, leading to ompF repression.
Recruitment of RNAP to OmpR-Dependent Promoters
OmpR-mediated transcription occurs through a recruitment mechanism whereby OmpR facilitates binding of RNAP at OmpR-dependent promoters. The ompF and ompC promoters contain –10 elements that deviate from the E. coli consensus sequence (see below). A genetic study examined the role of each –10 element in OmpR-dependent transcriptional activation.
-
Consensus TATAAT
ompF AAAGAT
ompC GAGAAT
When the first base of either –10 element was converted to a consensus T (labeled in red), the promoters were highly active in an ompR mutant background (127). In an ompR null background, the level of transcription was further enhanced. The nonconsensus –10 promoter element necessitates the requirement for OmpR in transcriptional activation. The addition of OmpR and/or mutation toward the consensus sequence results in stimulation of transcription. These results are consistent with a recruitment mechanism for RNAP binding to promoter DNA. OmpR recruits RNAP to promoter DNA via a direct protein-protein interaction, or alternatively, OmpR binding could drive a structural change in the promoter that then facilitates RNAP binding. Since a single nucleotide change can stimulate OmpR-independent transcription, the former possibility seems more likely. A point mutation is more likely to result in the formation of a favorable nucleotide-amino acid interaction, rather than mimicking a structural change in the DNA due to binding of an upstream activator.
The location of the transcription factor binding site determines the requirement for αCTD at the promoters under examination. OmpR binds to sites upstream of the –35 region of the ompC promoter and requires the reconstituted RNAP containing the full-length αCTD to activate transcription, suggesting that a direct interaction occurs between OmpR and αCTD (128). In contrast, the OmpR family member PhoB binds to a site at the pstS promoter in the position normally occupied by the –35 element and does not require αCTD; it directly interacts with the σ70 subunit of RNAP holoenzyme (103, 104). Thus, OmpR subfamily members can activate transcription via interactions with different subunits of RNAP, and their mechanism of activation is not conserved throughout the subfamily.
Subsequent genetic studies identified substitutions in the α subunit that affected expression uniquely at the porin genes but not at other test promoters (129, 130). However, it was not determined whether the substitutions affected the interaction between OmpR and α or the interaction between α and promoter DNA. It is well established that α stimulates transcription at certain promoters by directly binding to high-affinity sites termed UP elements (reviewed in reference 131), but most activator-dependent promoters do not contain UP elements. It has been proposed that at some promoters that lack UP elements, nonspecific or low-affinity interactions between α and DNA are stabilized by interactions with an activator and α or with σ70 and α (132–138). Thus, it remains possible that mutations in α that specifically affect transcription of OmpR-dependent promoters alter the interaction of α with DNA, and not with α and OmpR.
Evidence for an interaction between OmpR and α was provided by the identification of a suppressor mutation in rpoA (encoding the α subunit) of the OmpR mutant P179L (139). The suppressor resulted in a glycine substitution for valine at position 264 of α (139). Surprisingly, the cocrystal structure of a CAP-αCTD-DNA complex identified residue 264 of αCTD as making direct contacts with the DNA backbone (132). It thus seems more likely that the V264G substitution enhances the affinity of αCTD for DNA, rather than suppressing the OmpR-positive control defect. From further analysis of the OmpR P179L mutant, it is evident that the mutant is severely defective in binding to DNA. This defect can be overcome by increasing the concentration of the mutant in an in vitro transcription assay, leading to a stimulation of transcription (Walthers and Kenney, unpublished results). Thus, it is still unresolved whether the requirement for α at OmpR-dependent promoters is the result of an interaction with OmpR and α or the promoter DNA and α. Two studies reported inhibitory effects of overexpression of α on the ompF and ompC promoters, suggestive of a direct OmpR-α interaction (130, 140). High concentrations of α inhibit OmpR-dependent transcription of ompF, presumably by sequestering OmpR from the promoter. However, it is possible that α bound to promoter DNA prevents RNAP holoenzyme from interacting with OmpR and/or the promoter DNA, a scenario that would not require a direct interaction between OmpR and α.
OmpR RNAP Interaction Surface
The precise interaction surfaces on OmpR and α remain undetermined. Genetic screens for positive control mutants, defined as mutants defective in their interaction with RNAP but not with DNA, identified three potential surfaces on OmpR for interaction with RNAP (105, 141, 142). The criteria for positive control were narrowly defined (see below), and most mutants ended up having binding defects. This does not rule out a direct effect with α, but it does indicate that more experiments are needed to identify the interaction surface. These surfaces include the N-terminal receiver domain and two regions in the C terminus, the α loop and the α1-α2 loop (see Fig. 5). However, the lack of a DNA binding defect was confirmed only with the high-affinity site C1 (105), and binding at other loci was not examined. In two of these cases, a partial defect in binding to C1 DNA was apparent using a direct in vitro approach (141, 142). Altered binding was shown to occur at the low-affinity sites without a simultaneous defect at the high-affinity C1 site (98), suggesting that high-affinity binding is not a stringent enough requirement for a positive control mutant. This result raises the possibility that the positive control mutants previously identified could be defective in DNA binding at the low-affinity sites, rather than having an impaired interaction with RNAP. Because OmpR binding to C3 is required for activation of the wild-type ompC promoter, defects in binding to this region would confer a transcriptional activation defect (98).
A subset of the positive control mutants was expressed and purified to examine their DNA binding and transcriptional activation properties. OmpR R42H is a substitution in the N-terminal domain (105); OmpR G191S and A196V are substitutions in the so-called α loop (105), which is the turn of the HTH motif (94). OmpR P179L (105, 141) and S181P (142) are substitutions in the α1-α2 loop immediately upstream of the HTH motif. Four of the five mutants had DNA binding defects to varying degrees at both promoters (Walthers and Kenney, unpublished results). S181P was defective in binding to ompC but protected ompF from DNase I cleavage similarly to wild-type OmpR. All of the mutants, with the exception of S181P, were able to significantly activate transcription in vitro. The results indicate that the substitutions in the N terminus and the α loop are actually DNA binding mutants and not positive control mutants. When the DNA binding defect is overcome by increasing the concentration of the mutant proteins, they are able to activate transcription. Based on these results, it is proposed that a location in the N terminus containing R42 (α1) and the α loop are potential candidates for regions that determine DNA binding specificity (Walthers and Kenney, unpublished results). The α1-α2 loop remains a likely candidate as an RNAP interaction surface. The crystal structure of the OmpR C terminus (OmpRc) reveals that residues in this region interact with residues within the recognition helix (94, 102, 143). Thus, it is not unexpected that substitutions in the α1-α2 loop could also affect DNA binding. Clearly, additional studies are required to rigorously identify the regions of OmpR required for an interaction with RNAP.
Affinity Model of Porin Gene Regulation
A model that explained the differential regulation of ompF and ompC was referred to as the affinity hypothesis. The model was based on genetic analysis of a series of mutants that indicated that (i) OmpR∼P is the form that regulates the porin genes and OmpR does not play a role (4) and (ii) EnvZ controls the concentration of OmpR∼P via its biochemical activities (described in “EnvZ Biochemical Activities Underlie Signaling” above) (56). We now appreciate that OmpR plays a surprising, noncanonical role in regulating many genes (see “OmpR is a Global Regulator” below and “A New View of EnvZ/OmpR Based on Noncanonical Signaling”) (2, 3, 24). The hypothesis is outlined briefly as follows: at low osmolality, there is a low level of OmpR∼P present, due to either a low activity of the EnvZ kinase or a high activity of the EnvZ phosphatase. OmpR∼P binds to high-affinity sites on ompF that activate its expression. At high osmolality, the kinase activity of EnvZ increases, or the phosphatase activity decreases, increasing the OmpR∼P concentration. OmpR∼P binds to low-affinity sites on ompF that repress OmpF expression, and it also binds to low-affinity sites on ompC that activate expression of OmpC. The hypothesis had two basic predictions: that phosphorylation of OmpR increases its affinity for the porin regulatory regions and that the upstream sites have different affinities for OmpR∼P. The ompF activating sites should be high affinity, the ompF repressing site should be low affinity, and the ompC activating sites should be low affinity. Thus, at low OmpR∼P (low osmolality), the high-affinity site on ompF would be occupied, leading to OmpF production. At high OmpR∼P (high osmolality), the low-affinity sites at ompF would be bound, repressing ompF transcription, and the low-affinity sites at ompC would also be occupied, activating transcription of ompC. It was proposed that the differences in affinity would need to be on the order of 20-fold to account for the observed differences in OmpF and OmpC expression levels (56).
Testing the Affinity Model
A quantitative analysis of the binding affinities of OmpR and OmpR∼P for individual OmpR binding sites and the composite ompF and ompC promoters was performed using fluorescence anisotropy (106). OmpR was phosphorylated by AcP, and OmpR∼P concentrations were measured on a C4 column using reverse-phase high-pressure liquid chromatography. The binding assays revealed that OmpR∼P has similar affinities for both the isolated F1 and C1 sites. This result was surprising, because the affinity hypothesis had predicted that the F1 site should be of considerably higher affinity than the C1 site. When OmpR∼P binding to the composite F1-F2-F3 and C1-C2-C3 sites was examined, the affinity for ompF DNA (Kd = 15 μM) was only 2-fold higher than for ompC DNA (Kd = 31 μM). The binding assays indicated that the affinity of OmpR∼P for each promoter was not sufficiently dissimilar to account for osmoregulation of porin gene expression on the basis of differences in binding site affinity. More interestingly, the binding data indicated that the affinity of unphosphorylated OmpR was greater for ompC (Kd = 87 μM) than for ompF (Kd = 194 μM). Thus, even at low OmpR∼P concentrations, it is predicted that occupancy of ompC and ompF by either OmpR or OmpR∼P would be quite similar. Considering that the affinities of OmpR∼P for ompF and ompC were quite similar, the simplest explanation for the lack of ompC activation at low osmolality is a type of gene silencing mechanism. Such a mechanism would prevent OmpR and/or RNAP binding to negatively regulate steps downstream of RNAP recruitment (144). Occlusion of OmpR binding to ompC at low osmolality by another bound factor or by an altered DNA structure that requires high concentrations of OmpR∼P to activate ompC is in agreement with one aspect of the affinity hypothesis: that differences in the porin levels in response to changes in medium osmolality require higher concentrations of OmpR∼P.
Differential Regulation of the Porin Genes Is Driven by Conformational Changes in OmpR
The affinity hypothesis was elegant and simple, but if the hypothesis is no longer valid, how can differential osmoregulation of the porin genes be explained? Recent studies of the OmpR mutant T83I led to an alternate model for differential porin gene expression (98). A threonine near the phosphorylation site is a highly conserved residue among RRs (position 83 in OmpR). It participates in a phosphorylation-dependent conformational change in the N-terminal receiver domain and transmits it to the C-terminal DNA binding domain of OmpR. Upon phosphorylation at Asp55, the side-chain hydroxyl of T83 hydrogen binds to the phosphoryl group. This creates a pocket that enables Y102 to rotate from an outward, solvent-exposed to an inward, buried position (112, 145–148). This “aromatic switch” is likely a general mechanism to transduce the phosphorylation event into a conformational change in the protein. Such a conformational change would then modulate protein function, for example, by increasing affinity for DNA. When overexpressed in vivo, OmpR T83I was unable to activate transcription of either ompF or ompC. This defect was not due to a defect in phosphorylation by EnvZ. DNase I footprinting assays indicated that the OmpRT83I mutant was able to bind to ompF identically to wild-type OmpR yet displayed altered binding at ompC. Thus, OmpR must adopt different conformations when bound to ompF versus ompC. In the case of T83I, the mutant appears to be locked into a low osmolality conformation that enables normal binding at ompF but not at ompC. It is especially intriguing that this substitution is in the phosphorylation site, not in the DNA binding domain of OmpR. Binding at ompC would require that OmpR adopt a different conformation, and this switch is prevented by the substitution. The mechanism that facilitates the conformational change of OmpR when bound to one promoter versus the other remains elusive but will hopefully be revealed when cocrystal structures are available. It is possible that OmpR changes selectivity in response to intracellular changes in osmolality or pH (3). Another possibility is that the different conformation stabilizing DNA binding at ompC occurs post-initial DNA recognition (117). In other words, once OmpR is bound to ompC DNA, a conformational change might occur to facilitate oligomerization, RNAP interaction, or some other event required for transcriptional activation. A shift to high osmolality may directly or indirectly alter the structure of the ompC promoter that facilitates DNA binding. For example, a previous report demonstrated that integration host factor (IHF) protects approximately 40 bp upstream of the OmpR binding sites at the ompC promoter (149). If IHF binding at ompC depends on environmental osmolality, binding or release at high osmolality could lead to a structural change in the downstream DNA that favors OmpR binding. In agreement with this prediction, a study demonstrated that IHF binding to an upstream activating sequence of the ilvPG promoter mediates duplex destabilization in the –10 element that favors RNAP binding (150). In other words, a structural change in the DNA transmitted by distally bound IHF favors RNAP binding in the absence of any direct interaction between the two proteins. The characterization of OmpR T83I revealed an additional property of OmpR-dependent transcriptional activation. T83I binds to ompF similarly to wild-type OmpR, yet it is unable to activate transcription. Since EnvZ can phosphorylate T83I in vitro, the substitution likely confers a defect in signaling that is subsequent to phosphorylation and is required for transcriptional activation (98). Likely possibilities are OmpR oligomerization or a conformational change that is required for a productive interaction with RNAP.
The OmpR mutant V203M constitutively expresses ompF but is defective at activating ompC. V203M has a reduced affinity for ompC DNA compared to wild-type OmpR (151). In a himA null background (encoding one of the two IHF subunits), the ompC expression defect was suppressed (149). These results suggest that IHF inhibits OmpR binding at the ompC promoter, although this possibility has not been tested directly. It is unclear whether or not IHF levels are coordinated to changes in medium osmolality and under what conditions IHF binds at ompF and ompC (i.e., at low versus high osmolality or both). Thus, the precise contributions made by IHF to differential porin gene expression and the signals that feed into this pathway remain elusive. A further complication is that IHF has also been shown to negatively regulate transcription of the ompR-envZ locus in both in vitro and in vivo assays (152). An intriguing question is whether or not changes in IHF levels and/or function in response to changes in medium osmolality are important for differential regulation of the three loci. However, IHF is a pleiotropic, global regulatory protein, and any models concerning IHF-dependent regulation will likely be difficult to test in vivo.
OmpR is a Global Regulator
OmpR is well known for its role in modulating the ratio of OmpF to OmpC in the cell in response to changes in osmolality, but it is now evident that OmpR plays a much wider role in regulating gene expression. Earlier transcriptome analysis reported that an ompR/envZ deletion resulted in changes in the expression of 125 different genes (153). The ompR/envZ response was second only to a deletion of the arcA/B two-component system in terms of the number of genes affected. Although some of these changes may be the result of defects in porin synthesis, they include a diverse array of genes, such as those necessary for flagellar-mediated motility, amino acid biosynthesis (cysteine and leucine), and enterochelin synthesis. The lack of both EnvZ and OmpR results in a variety of phenotypic changes, including increased hexose utilization (allose, fructose, mannitol, N-acetyl-d-glucosamine, and glucose), increased resistance to antibiotics (cephalosporins, β-lactams, topoisomerase inhibitors, and folate antagonists), and decreased resistance to some metal ions (cobalt, selenite, chromate, lithium, and dichromate) but, interestingly, little change in sensitivity to osmolality (154). More recently, a deletion in ompR was shown to increase expression of a variety of drug exporters (155). Previous work from many groups has shown that OmpR plays a role in the regulation of a wide variety of genes, including the flagellar master regulon flhDC in E. coli, but not in Salmonella (156), expression of the csgDEFG operon and curli fimbriae production (157–159), expression of cryptic porins OmpS1 and OmpS2 in Salmonella enterica serovar Typhi (160, 161), the stationary-phase acid tolerance response in S. enterica serovar Typhimurium (162, 163), and expression of many virulence genes (110, 111, 164–167).
In E. coli MG1655, whole-genome expression profiling identified acid-responsive genes including chaperones, regulators, genes involved in metabolism (e.g., glutamine decarboxylase), and some genes associated with the cell envelope (168). In another study, the E. coli response to mild and strong acidic conditions was compared, revealing a complex transcriptional program that was dependent on OmpR and the switch between aerobic and anaerobic growth (109). OmpR was connected to genes involved in pyruvate metabolism and glycolysis, signal transduction and transport, and some components of the glutamate decarboxylation system. Direct OmpR targets were not identified, most likely because OmpR regulation of acid stress pathways occurs usually via repression of transcription, which has less precise recognition requirements (3, 24). Identifying OmpR binding sites by sequence gazing is difficult, because OmpR has a high capacity for nonspecific binding to DNA, in part because it makes more phosphate backbone contacts and fewer DNA base contacts (95). Thus, it is now evident that OmpR drives a major reprogramming in response to acid and osmotic stress. What remains to be determined is how many of these genes are directly regulated by OmpR.
OmpR also regulates two small RNAs, omrA and omrB, which negatively regulate the iron transport genes cirA, fecA, and fepA and the outer membrane protease ompT (169). DNaseI footprinting determined that omrA/B binding sites were exquisitely sensitive to OmpR∼P (binding at nM concentrations), and unphosphorylated OmpR did not bind to the promoter DNA (Walthers and Kenney, unpublished results). This observation was validated by in vitro transcription assays that were also completely dependent on OmpR∼P (and not OmpR), although the precise concentration of OmpR∼P in the reaction was not determined (107).
A larger role for OmpR in regulating gene expression is particularly evident in the response to acid and osmotic stress in S. Typhimurium and E. coli (2, 3, 24, 109, 170). Based on an observed intracellular pH threshold of ∼6.5, OmpR represses distinct sets of genes that neutralize the bacterial cytoplasm, thus enabling acidification (3). During acid stress in S. Typhimurium and E. coli, OmpR contributes to cytoplasmic acidification by repressing the cadC/BA operon (3, 24). CadC is in the OmpR subfamily of response regulators, and normally it activates transcription of cadBA. CadA is a lysine decarboxylase, which consumes a proton during decarboxylation. The product, cadaverine, is then transported out of the cell by the CadB antiporter. Repression of cadC/BA by OmpR thus prevents neutralization. Because the pH optima of the CAD system is 6.1 to 6.5 (171), it is the most important acid stress system at vacuolar pH (24), and shutting it off enables the bacterial cytoplasm to acidify in the vacuole.
OmpR also promotes cytoplasmic acidification in response to osmotic stress, but different pathways are involved. In S. Typhimurium, OmpR represses the alternative stationary phase sigma factor, rpoS, relieving RpoS repression of yghA. YghA is a putative oxidoreductase that is predicted to produce protons (3). In E. coli, the intracellular pH was less acidic and OmpR regulated different pathways. OmpR repressed speF, the arginine decarboxylation system, which has a higher pH optimum of 7.0 (172). Normally, ornithine decarboxylase decarboxylates arginine, consumes protons, and produces putrescine, which allows for recovery from acidification. Repression of speF by OmpR prevents recovery at high osmolality (3).
A recent microarray analysis compared the OmpR-dependent response to acid stress in E. coli and reported that about six times as many genes were involved compared to S. Typhimurium (2). The involvement of 1,360 genes in the OmpR-dependent E. coli MG1655 acid stress response (and 1,538 genes in the overall OmpR-independent acid stress response) was similar to a study of E. coli BW25113, in which 1,871 genes were differentially expressed after a 15-minute acid exposure to pH 5.5 (109). These two studies differ from a ChIP-on-chip study using the E. coli K-12 strain CSH-50 (170). In that study, only 144 OmpR-regulated acid stress genes were identified in CSH-50, which was only ∼10% compared to the 1,360 genes identified in E. coli MG1655. In contrast, the magnitude of the response of S. Typhimurium was similar: 240 versus 212 acid stress genes identified (2, 170). In E. coli CSH-50 and S. Typhimurium, 15 OmpR-regulated genes were common (170), whereas in E. coli MG1655 and S. Typhimurium strain 14028, 25 genes were common (2). Surprisingly, there was very little overlap between the genes involved in both acid and osmotic stress pathways of E. coli and S. Typhimurium (2, 170). The genes that were in common were known OmpR targets: ompC, ompF, and the tripartite permease, tppB. These same targets appear in the common genes of the acid and osmotic stress responses of S. Typhimurium and E. coli (2). Extensive genetic differences between E. coli K-12 CSH-50 (173, 174) and MG1655 likely explain the poor overlap between genes identified in these studies.
Flagellar Biosynthesis
The flagellar master regulator FlhDC is encoded by the flhDC operon and is responsible for activating the expression of motility and chemotaxis genes, including the flagellar components, the motor apparatus, the chemoreceptors, and che genes (for a review, see references 175 and 176). Several studies report that expression of the flhDC operon was partially affected by AcP concentration (177–179). Shin and Park demonstrated that OmpR∼P binds to the promoter and represses flhDC (156). Mutations in phosphotransacetylase (pta) or ompR increase flhDC expression, whereas mutations in acetate kinase (ackA), envZ11, or increasing osmolality decrease flhDC expression. On soft-agar motility plates, Δ(ompR–envZ) mutations have a stronger swarm phenotype than the wild type (153). Therefore, AcP may regulate flagellar biosynthesis through phosphorylation of OmpR, which binds to the flhDC promoter region and inhibits expression of the FlhDC master regulator. Electrophoretic mobility shift assays and DNase I footprinting identified two binding sites in the flhDC promoter region (156). Phosphorylation of OmpR leads to a 10-fold increase in DNA binding affinity, as also observed for binding at ompF and ompC (106), and results in protection at regions –179 to –116 and –17 to +49 from the flhDC transcriptional start site. An IHF binding site is located between the two OmpR binding sites at flhDC. A repression loop, similar to that predicted at ompF may also occur between the two binding sites at flhDC to inhibit transcription of this operon.
Curli Fimbriae Production
Many bacteria form biofilms on surfaces, and life in a biofilm is a prevalent lifestyle for microbes in the environment. A biofilm is characterized as a community of microorganisms suspended in a polysaccharide matrix (180). Bacteria in these environments are extremely resistant to most antimicrobial agents and display different patterns of gene expression and morphology distinct from their free-swimming counterparts. A mutation in ompR (Leu-43 to Arg substitution; ompR234 allele) causes a dramatic increase in bacterial surface binding and biofilm formation compared to the wild type (159). This phenotype is caused by an overproduction of curli, which are long fimbrial structures composed of the products of the csgBA operon. In either wild-type or ompR null backgrounds, curli production is substantially reduced. Expression of csgBA is stimulated by the transcriptional activator CsgD, whose expression is dependent upon OmpR (158). Although OmpR is proposed to act as a positive regulator at csgD, curli formation is repressed at high osmolality. S. Typhimurium contains six OmpR binding sites upstream of the csgD transcriptional start site, one of which is reported to have an inhibitory effect on csgD expression (157). In Salmonella, OmpR also regulates the SsrA/B two-component system (110, 111), and the RR SsrB noncanonically activates csgD in the absence of phosphorylation by derepressing H-NS (181–183).
Virulence
OmpR modulates the virulence properties of several pathogens, and OmpR mutants have a reduced ability to infect hosts. In Shigella flexneri, OmpR alters expression of the vir genes necessary for invasion of epithelial cells (164). Expression of the vir genes is reduced in an envZ null mutant but can be restored by increasing medium osmolality. In an ompR-envZ double mutant, osmolality changes are uncoupled from vir expression, and the mutants are attenuated in the invasion of HeLa cells. In Yersinia enterocolitica, an ompR null mutant is essentially avirulent and is unable to infect orally challenged BALB/c mice beyond 21 days postinoculation (166). The ompR null strain exhibits decreased survival when exposed to high osmolality (1.5 M NaCl) or high temperature (55°C) compared to the wild type. It remains to be determined whether the avirulent phenotype of Y. enterocolitica ompR null strains is caused by this increased susceptibility to environmental stress or, more likely, whether OmpR directly interacts with the regulatory regions of a subset of virulence genes necessary to sustain infection.
In S. Typhimurium, ompR and envZ deletions are attenuated for virulence and are unable to kill phagocytes and spread to other macrophages. The avirulent phenotype is not a result of reduced expression of ompF, ompC, or tppB (167). OmpR activates expression of SsrAB, a two-component regulatory system located on Salmonella pathogenicity island 2 (110, 111). SsrA/B subsequently regulates a type III secretion system that is required for Salmonella to replicate inside macrophages. This is one of only a few examples where a response regulator of one two-component system affects the transcription of another two-component system. Footprinting analysis identified several OmpR binding sites upstream of ssrA and both upstream and within the ssrB coding sequence (110, 111). The expression of the kinase SsrA is uncoupled from the expression of the response regulator SsrB (89, 184). This is unusual compared to most two-component regulatory systems in which the ratio of kinase to response regulator is tightly controlled. In S. Typhi, OmpR also directly binds and regulates the Vi capsule regulator tviA (185).
A Caution Regarding C-Terminal Fusions to OmpR
Recent ChIP-on-chip results with OmpR reported OmpR binding at the mgtC promoter (170). In contrast, microarray and real time quantitative reverse transcription PCR (qRT-PCR) analysis of OmpR-regulated genes at acid pH did not identify mgtC as a target of OmpR regulation, nor did mgtC contribute to intracellular acidification (24). A likely explanation for this discrepancy is that OmpR containing a 3XFLAG tag was employed in the ChIP-on-chip study, which also contained a D55E substitution (170). C-terminal tags to OmpR affect its DNA binding ability (185) and also alter its specificity (186). For example, the D55E-3XFLAG-tagged OmpR required concentrations of >1 μM to bind in electrophoretic mobility shift assays (187), even though the affinity of wild-type, unphosphorylated OmpR for the porin genes ompF and ompC is ∼150 nM (106). OmpR targets identified by SELEX (186) were not able to be validated (Y. Gao and L. J. Kenney, unpublished observations), and different OmpR targets were identified depending on whether a C-terminal or N-terminal tag was employed (186). Thus, extreme caution should be used when interpreting results using OmpR C-terminal fusions.
A NEW VIEW OF EnvZ/OmpR BASED ON NONCANONICAL SIGNALING
The old view of two-component signaling was based on a phosphorelay in which the membrane-embedded HK was activated by an external signal (Fig. 3). An environmental signal results in a conformational change that drives ATP binding and subsequent autophosphorylation at the conserved histidine (His243 of EnvZ). The RR then engages the HK∼P, and phosphotransfer to the RR occurs. The RR∼P is then released from the HK to find its target DNA binding sites, allowing activation or repression of transcription. In some systems, the HK plays an additional role as a phosphatase to turn over the RR∼P, but in others, this step is controversial (see above) (67).
Our new view of signaling is based on our recent observations of noncanonical signaling by EnvZ/OmpR in response to acid pH (2, 3, 24) and the HDXMS readout of EnvZ conformational changes in response to osmolytes (1, 32). During noncanonical signaling, EnvZ phosphorylation is very low (for example, at acid pH [3]), but OmpR still binds to EnvZ and dimerizes, due to an effect of acid pH on the OmpR binding site of EnvZ (the SINKDIEE peptide). OmpR dimerization mimics an activated state that then binds to DNA and activates or represses transcription (see reference 181 for a review). This signaling is noncanonical in that the reaction does not involve OmpR phosphorylation but requires an interaction of OmpR with EnvZ (3).
The prominent changes in EnvZ conformation as determined by HDXMS after exposure to osmolytes involved two loci: the histidine-containing peptide (HDXMS peptide amino acids 238 to 254) and the OmpR binding site (HDXMS peptide amino acids 269 to 276: SINKDIEE). Note that the actual region might be larger than the peptides identified by HDXMS; for example, C277 is C-terminal to the SINKDIEE peptide, and fluorescent labeling of C277 eliminates OmpR interaction with EnvZ (21). In the His243 locus, the central positioning role of the flanking carbonyl and acidic side chains also make the His-Asp/Glu dyad sensitive to acid pH sensing, although only osmolyte-induced conformational changes enhance autophosphorylation. Lower pH, on the other hand, increases protonation of Asp244, and coupled with stronger helical main chain bonding would alter conformation of the His243 locus without phosphorylation.
Analysis of the side chain interactions during EnvZ activation provides us with a view in which the signal (intracellular acid or osmotic stress in the case of EnvZ) modifies in parallel both the RR binding site and the histidine-containing peptide of the HK. This change drives a switch between a nonfunctional interaction of the RR with the HK to a functional interaction, promoting autophosphorylation and subsequent stimulation of phosphotransfer to the RR. In this scenario, autophosphorylation and phosphotransfer are essentially coupled because the RR is already complexed to the HK. The phosphorylated RR∼P dimer (or the unphosphorylated RR dimer in the case of noncanonical signaling) binds DNA, or may already be bound to DNA (117), and activates or represses transcription. The revised series of events is illustrated in Fig. 4.
“An Asp for an Asp” Drives Phosphotransfer
In the resting state, OmpR is bound to the region spanning the EnvZ SINKDIEE locus via interactions in the α1 helix of the OmpR N-terminal receiver domain (64). In step 1, a signal activates ATP binding and stimulates the acidic cluster of the SINKDIEE locus, loosening its interaction with OmpR at its α1 helix. The His243-Asp244 dyad of EnvZ is destabilized via osmolytes or acid pH (32, 188), reducing the brake that keeps the histidine-spanning peptide disordered. The helix containing His243 forms, allowing changes in the histidine side chain rotamer, which promote phosphorylation of His243 (1). Subsequently, Asp55 and residues in the β3-α3 and β4-α4 loops of the OmpR N terminus engage the His∼P of EnvZ, and phosphotransfer to OmpR occurs. As a consequence, the inhibitory His-anchoring role of Asp244 in EnvZ is now replaced by Asp55 of OmpR. Thus, an Asp244(EnvZ)-Asp55(OmpR) switch drives phosphotransfer.
EnvZ and OmpR thus jointly function as a composite signaling entity, wherein changes due to osmolality are detected in a continuum by modular units in both proteins, leading to autophosphorylation and phosphotransfer. This new model incorporates inherent specificity that is built into the unique interface mediated by the HK bound to its RR (SINKDIEE locus, OmpR binding site of EnvZ). This programs each HK-RR cognate complex to respond with exquisite specificity to precise environmental stimuli while retaining modular elements of ATP binding, autophosphorylation, and phosphotransfer chemistry, which are conserved across nearly all two-component signaling systems. This modularity also effectively allows for noncanonical signaling inputs that can modulate the HK-RR interaction interface to drive additional signaling responses. For example, an altered conformation of OmpR involves a switch from α1 engagement with EnvZ in the resting state to engagement via β3-α3 and β4-α4 loops as the EnvZ SINKDIEE peptide changes conformation in acidic pH. This generates an altered output response, e.g., activation of the acid stress response (2, 3, 24), and explains how the HK-RR composite signaling unit can be specifically driven by phosphorylation-independent noncanonical signaling outputs.
How Do Signals Alter EnvZ/OmpR Conformation?
A Combination of Side Chain and Peptide Backbone Effects Drives Signaling
The structures of diverse HKs indicate that the autophosphorylated His locus is always positioned within α-helices. In addition, directly C-terminal to the His residue is an acidic Asp/Glu residue that is strikingly conserved in almost all HKs, except PhoQ (32). A series of interactions relays multiple signaling inputs that converge onto the His-Asp/Glu dyad to alter the signaling state of the His and drive autophosphorylation. Parallel side chain interactions in tandem with helical backbone contacts mediate a coordinated response to osmolytes or acid pH in the case of EnvZ.
Side Chain Effects from the His-Asp Dyad
Specific side chain interactions at the His-Asp dyad directly impact the stability of the helical backbone holding the His side chain. These side chain interactions then modulate the stability of the helical backbone (Fig. 8). The positioning of His and Asp residues and their effects on the stability of α-helices has been extensively examined by Huyghues-Despointes and Baldwin (189). The occurrence of His and Asp in helices is not stochastic. Positioning of an Asp residue C-terminal to a His residue enhances α-helix stability only when the Asp is in the i, i+3 and i, i+4 positions. In EnvZ, the Asp is in the i, i+1 position. This makes the His-Asp dyad in EnvZ intrinsically destabilizing, and it works to maintain the histidine locus in a locally disordered state (32). Because the Asp is at an i, i+1 position, generation of more favorable side chain interactions necessitates puckering of the backbone helix at His243. This forms the core of the signaling unit: stabilization of the helical backbone occurs in EnvZ in the presence of osmolytes and negatively impacts the His-Asp side chain contacts. Thus, osmolytes shield the His-Asp side chain contacts, which promotes the ordering of the helical backbone and repositions the His243 rotamer. These events stimulate autophosphorylation. Interestingly, osmolyte-mediated stabilization has been observed only in the His-Asp orientation and not in the Asp-His orientation of synthetic peptides (189).
Figure 8.
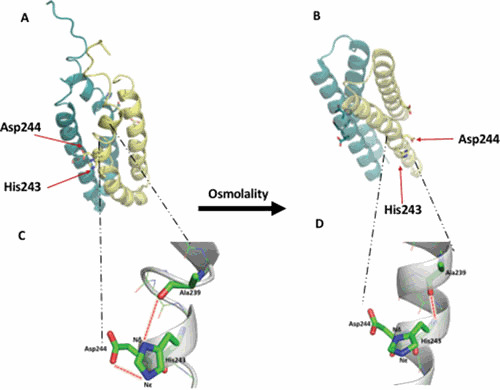
Model of EnvZ osmosensing and autophosphorylation. High-resolution structures of the His243 neighborhood under conditions of low osmolality (by NMR PDB ID:1JOY) (A) and high osmolality (by X-ray crystallography PDB ID: 4KP4). (B) The osmolality-dependent conformational change is associated with helix stabilization across the four-helix bundle subdomain. (C) An inset shows the local disorder within the His243-containing helix. The imidazole ring nitrogens (blue) are anchored by competing H-bonds with the Ala239 backbone carbonyl group and Asp244 side chain (red dashed lines) to maintain low basal levels of His243 phsophorylation. (D) Osmolality-induced helical backbone stabilization strengthens the backbone H-bond between the Ala239 carbonyl and aids in positioning the Asp244 side chain for enhanced His243 phosphorylation at Nε.
Peptide Backbone Stabilization
Examination of the dynamics of the cytoplasmic domain of EnvZ by HDXMS revealed that the four-helix bundle formed the primary signaling locus for osmosensing (1). Residues flanking His243 were positioned in an island of greater disorder relative to the rest of the protein (20). Osmolytes facilitated stabilization of this locus, resulting in enhanced autophosphorylation through integration of helical main chain H-bonding and side chain interactions (32). A recent structure of EnvZc solved by X-ray crystallography at high osmolality further corroborated these results (190) (Fig. 8). The “side chain-main chain swap” model we developed (1, 32) emerged from the use of strategic inhibitory and activating mutants in EnvZ that were located one turn above and one turn below the phosphorylated His243 that had opposing effects on autophosphorylation (51, 188, 191).
The Effect of Osmolytes or Acid pH on EnvZ: a Two-Way Street
Osmolytes function to couple helical backbone stabilization (1) and relieve the His-Asp dyad side chain interactions. Stabilization occurs when the H-bond between the carbonyl C=O of Ala239 and the imidazole nitrogen of His243 switches to that between the carbonyl C=O of Ala239 and the main chain amide N-H of His243 (32), freeing the Nε for phosphorylation. Osmolytes also shield His243-Asp244 side-chain interactions, releasing the brake and promoting stabilization of the helical backbone. These two effects of osmolytes are coupled: altering the His-Asp side chain interactions promotes helical stabilization of the His243 backbone, and helical backbone stabilization at the His243 locus alters the positioning of the His-Asp dyad. Together, these parallel effects of osmolytes on helical backbone and side chain interactions constitute the basis of osmosensing and enhanced His243 phosphorylation.
Substitution of Asp244 with Ala in EnvZc eliminated autophosphorylation of His243 (192) by enhancing ATP turnover (32). The Asp244 side chain serves as a brake on His243 autophosphorylation at low osmolality, but repositioning of Asp244 in the activated conformation also stabilizes phospho-His243. Thus, in the phosphorylation experiments (192), no His∼P was detected, because its turnover was enhanced. Both of these results point to a critical role of the Asp/Glu residue of the His-Asp dyad in modulating both His autophosphorylation and stability of the phospho-histidine in HKs.
The His-Asp dyad also forms the integrative node for relaying the effects of RR interactions. In EnvZ/OmpR, OmpR contact with EnvZ through the OmpR α1 helix is relayed to the acidic SINKDIEE cluster in EnvZ through a network of charged residues that modulate the conformation of the His-Asp dyad. The composite EnvZ/OmpR interface forms the basis for specificity of two-component signaling responses while retaining the conserved chemistries of His-Asp phosphorelays. The His-Asp dyad is highly sensitive to protonation states and pH, providing an alternate stimulus for mediating a phosphorylation-independent response in HK-RRs (2, 3).
If Nearly All HKs Possess the His-Asp Dyad, What Provides Specificity in the Signaling Response?
The answer to this question lies in the RR binding domain of the HK (SINKDIEE in EnvZ), which is unique for each HK-RR cognate pair. In an analysis of covarying residues between HKs and their cognate RRs, two apparent clusters of specificity determinants were identified in each HK and RR (64). In the HK, one cluster was at the base of the four-helix bundle and was implicated in NMR studies as a site of RR interaction (20); in EnvZ, this site was confirmed to be a site of interaction with OmpR by native SDS-PAGE and fluorescence cross-correlation spectroscopy (3, 21, 32). Osmotic stimuli affect the His-Asp dyad by releasing the brake and activating the helix surrounding His243 and also by driving the acidic SINKDIEE locus into an active configuration. The change in the SINKDIEE locus, which is coupled to activation by environmental stimuli, shifts the interface of OmpR interaction with EnvZ from the α1 helix of the OmpR receiver domain to the β3-α3 and β4-α4 loops of OmpR where Asp55 resides. Thus, OmpR binds to EnvZ using a different interface in its inactive and active signaling states, and this difference explains why OmpR∼P has a lower affinity for EnvZ than OmpR (66), because it has a different interface. This may also explain why AcP phosphorylation of RRs in vivo is reduced in the presence of the cognate HK, because the free RR concentration is reduced. Signal fidelity then resides in how signals affect the RR binding site of the HK; for example, noncognate HKs will not experience a change in their RR binding site in response to “EnvZ signals.” With advances in super-resolution imaging, we are optimistic that even low-abundance complexes such as EnvZ/OmpR can soon be visualized in single bacterial cells to examine how the complex is affected by environmental stimuli (see “Noncanonical Activation of OmpR” below).
Cytoplasmic Signaling
Despite evidence for the modular nature of the cytoplasmic domain of EnvZ, a stand-alone role for EnvZc was unequivocally demonstrated only recently (1). In this study, EnvZ lacking the N-terminal 179 residues, or EnvZc, fully restored osmosensing in an E. coli envZ null mutant, as well as osmolyte stimulation of His243 autophosphorylation. This result also clearly demonstrated that the TM and periplasmic domains were not obligatory for EnvZ function.
Noncanonical Activation of OmpR
Although unphosphorylated OmpR does not play a role in porin gene regulation (4), it plays an important role in regulating many genes in response to acid and osmotic stress. Earlier studies of the acid stress response reported a central role for OmpR (109), although no direct targets were identified. It was subsequently determined that the cytoplasm of E. coli and Salmonella became acidified in an OmpR-dependent manner in response to acid or osmotic stress (3, 24). During acid stress, OmpR repressed cadC/BA, preventing a return to neutral pH, by preventing expression of the CAD lysine decarboxylation system. Normally, in response to acid stress, the OmpR family member CadC activates cadB/A. CadA is a lysine decarboxylase, which consumes a proton during decarboxylation. The product of decarboxylation, cadaverine, is transported out of the cell by CadB. Repression of the CAD system by OmpR maintained an acidic cytoplasm (3, 24). Mechanistically, how does this work? From our unpublished studies, OmpR phosphorylation by AcP was lowest at pH 6.0 and highest at pH 8.0. EnvZ phosphorylation was also substantially reduced at acid pH to 28% of the level at pH 7.5 (3). The histidine nitrogen that is phosphorylated in HKs needs to be unprotonated to act as a nucleophile toward ATP (193). An aspartic acid mutant of OmpR behaved similarly to the wild-type OmpR in supporting acidification, and thus, phosphorylation was not required. Surprisingly, OmpR still required the EnvZ kinase (3), because an EnvZ mutant that was unable to interact with OmpR (21) was unable to support OmpR-dependent acidification (3). Thus, OmpR can act noncanonically to drive acid and osmotic stress responses. The observation that OmpR required an interaction with EnvZ (but not phosphorylation) highlights the importance of revising our thinking to consider HK-RR complexes in signaling, even though phosphorylation might not result from the interaction. In fact, recent evidence of noncanonical regulation by other RRs has been reported, including ArcA, RcsB, and others (see reference 181 for a recent review).
Analysis of OmpR in Single Cells
Due to the small size of bacterial cells and the diffraction limit of light, conventional fluorescence microscopy is of limited use (194). Super-resolution microscopy allows visualization of biological molecules far below the light diffraction limit (195). To image and count OmpR molecules in single cells, we constructed a photoactivatable fusion protein that was chromosomally expressed under its native promoter, and it was capable of activating ompF and ompC (21). We applied single-molecule localization microscopy to visualize OmpR molecules. The bacterial nucleoid was labeled with EdU (5-Ethynyl-2′-deoxyuridine) followed by click chemistry fluorescent labeling, and then a sequential workflow for imaging was applied (39, 90, 196). In single-molecule localization microscopy, the fluorescence signal is separated in time, and only a subset of fluorophores is detected, and their position can be precisely determined. Photoactivatable localization microscopy (PALM) can then provide copy numbers of OmpR in single cells (39, 90, 197), and single particle tracking (Spt-PALM) can determine the fraction of OmpR molecules that are immobilized (39, 89). At high osmolality, OmpR molecules were localized close to the edge of the nucleoid (Fig. 9), and the chromosome was not decorated by OmpR (39). At low osmolality, the nucleoid was more condensed than at high osmolality. In acid conditions, OmpR localized near the plasma membrane and appeared to be more clustered, consistent with a separate study in which OmpR dimerization and EnvZ interaction were acid-induced (3). Again, the nucleoids were more condensed (39, 89, 197). This finding is at odds with a report that argued that the nucleoids were more relaxed at acid pH, allowing OmpR to bind to more target genes (187). Single cells in acid were also ∼2 μm smaller than in LB (39).
Figure 9.
Sequential super-resolution imaging of OmpR-PAmCherry (red), bacterial cell membrane (green) and DNA (cyan) in various growth media: LB (A), 100 mM Tris pH 7.2 (B), 100 mM MES pH 5.6 (C), and 0.5× M9 buffer (D). Evidence for DNA compaction is evident in panels in the third row, where the green outlines the nucleoid edges. OmpR was distributed around the nucleoid edges (outlined in green) in LB, whereas it was more uniformly distributed in Tris buffer. In acidic (MES, pH 5.6) and hypotonic (0.5× M9) conditions, OmpR was recruited to the plasma membrane (scale bar = 1 μm). The number of images used for averaging cell length was 19 cells length 3.75 to 4.25 μm (A), 20 cells length 2.0 to 2.5 μm (B), 15 cells length 1.75 to 2.25 μm (C), and 13 cells length 1.5 to 2.0 μm (D). Reprinted from reference 39 under the terms of the Creative Commons CC BY license.
Early attempts to localize OmpR in cells employed a fusion protein in which the C terminus of OmpR was directly fused to green fluorescent protein (GFP), and the spatial distribution of OmpR-GFP was examined (194). Although the fusion was described as functional, the activation of the target gene ompC was only 20% compared to wild-type OmpR. This level was similar to an early photoactivatable mCherry (PAmCherry) fusion that we constructed (39). Addition of a linker between the C terminus of OmpR and the fluorescent protein increased activation to >70%. Fluorescent OmpR-GFP foci were visualized at what appeared to be near the membrane. The number of foci increased when EnvZ was overexpressed, and the foci were eliminated by overexpression of native OmpR; these foci were therefore interpreted as being complexes of OmpR-GFP with EnvZ (194). We were able to visualize EnvZ using PALM super-resolution imaging, although it required overexpression of EnvZ (21). We are optimistic that new advances in fluorophores and labeling (198) will now enable visualization of EnvZ/OmpR complexes in vivo, and these experiments are currently in progress in our laboratory.
When plasmids containing clusters of OmpR binding sites were imaged, additional OmpR-GFP foci were observed, indicative of OmpR binding to DNA (194). When OmpR was bound to these multiple DNA sites, OmpR-GFP foci near the membrane (suggestive of EnvZ/OmpR complexes) were not observed (194). The results from this study are supportive of a model in which EnvZ is first complexed with OmpR (although whether EnvZ is first phosphorylated or EnvZ is phosphorylated while complexed with OmpR is not known), and OmpR is then phosphorylated and released to bind to DNA (see Fig. 3). An improvement on this approach employed arrays of tetO and lacO binding sites near the ompF and ompC promoters, respectively (199). OmpR-yellow fluorescent protein colocalized with TetR-mCherry or cyan fluorescent protein-LacI and OmpR-yellow fluorescent protein foci were absent in the absence of envZ. The authors interpreted this to mean that OmpR was not phosphorylated and thus did not bind to DNA. However, the Kd of unphosphorylated OmpR binding to ompF and ompC is quite low, ∼100 to 150 nM (106). Based on our recent studies of noncanonical effects of OmpR, we offer a different interpretation of this result. We observed that OmpR was required to physically interact with EnvZ to activate the acid stress pathway (3), although OmpR phosphorylation was not required (reviewed in reference 2). In the presence of an envZ mutant, OmpR fails to dimerize, and no OmpR transcriptional effects would be observed. A similar effect might better explain the absence of OmpR binding to DNA in the envZ null strain (199). Advances in imaging should enable visualization of EnvZ/OmpR at higher resolution and may distinguish between EnvZ/OmpR complexes and OmpR/DNA complexes.
SUMMARY
Our recent work establishes the following events in EnvZ/OmpR signaling: OmpR is complexed with EnvZ in vivo. Osmolytes destabilize a negative effect of the His243-Asp244 dyad on the His243 locus, and this stabilizes the helix to better position the histidine side chain for enhanced phosphorylation. In concert, osmolytes also alter the conformation of the OmpR binding site, shifting the OmpR-EnvZ interface from the α1 interface of OmpR to the β3-α3 and β4-α4 loops, driving phosphotransfer to Asp55 of OmpR. Even though the features of the His locus and the His-Asp dyad are conserved in two-component systems, specificity to environmental signals resides in how the signals alter the RR binding site. Future work will focus on imaging these complexes in vivo using enhanced super-resolution techniques.
ACKNOWLDEGMENTS
We thank Melanie Lee and Diego Pitta de Araujo, MBI SciComm, Jeremy, Loo Chien Wang, Nikhil Kumar Tulsian, Department of Biological Sciences, NUS, for Figs. 2, 8, and Dasvit Shetty for Fig. 5A.
This work was supported by NIH grants AI123645, VAIOBX-000372, and MOE2018-T2-1-038 to L.J.K. and a Research Centre of Excellence grant in mechanobiology.
Contributor Information
Linda J. Kenney, Mechanobiology Institute, T-Lab, National University of Singapore, Singapore Department of Biochemistry & Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555.
Ganesh S. Anand, Department of Biological Sciences, National University of Singapore, Singapore
James M. Slauch, The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL
REFERENCES
- 1.Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. 2012. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J 31:2648–2659 10.1038/emboj.2012.99. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty S, Kenney LJ. 2018. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front Microbiol 9:2656 10.3389/fmicb.2018.02656. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty S, Winardhi RS, Morgan LK, Yan J, Kenney LJ. 2017. Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat Commun 8:1587 10.1038/s41467-017-02030-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slauch JM, Silhavy TJ. 1989. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol 210:281–292 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- 5.Slauch JM, Garrett S, Jackson DE, Silhavy TJ. 1988. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol 170:439–441 10.1128/jb.170.1.439-441.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wurtzel ET, Chou MY, Inouye M. 1982. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem 257:13685–13691. [PubMed] [Google Scholar]
- 7.Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277:24155–24161 10.1074/jbc.M110715200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Heyde M, Portalier R. 1987. Regulation of major outer membrane porin proteins of Escherichia coli K 12 by pH. Mol Gen Genet 208:511–517 10.1007/BF00328148. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Sato M, Machida K, Arikado E, Saito H, Kakegawa T, Kobayashi H. 2000. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl Environ Microbiol 66:943–947 10.1128/AEM.66.3.943-947.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas AD, Booth IR. 1992. The regulation of expression of the porin gene ompC by acid pH. J Gen Microbiol 138:1829–1835 10.1099/00221287-138-9-1829. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Todt JC, McGroarty EJ. 1992. Acid pH decreases OmpF and OmpC channel size in vivo. Biochem Biophys Res Commun 189:1498–1502 10.1016/0006-291X(92)90244-F. [DOI] [PubMed] [Google Scholar]
- 12.Dedieu L, Pagès JM, Bolla JM. 2008. The omp50 gene is transcriptionally controlled by a temperature-dependent mechanism conserved among thermophilic Campylobacter species. Res Microbiol 159:270–278 10.1016/j.resmic.2008.03.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Alphen WV, Lugtenberg B. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol 131:623–630 10.1128/jb.131.2.623-630.1977. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernando D, Kumar A. 2012. Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J Antimicrob Chemother 67:569–572 10.1093/jac/dkr519. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Lång H, Palva ET. 1993. The ompS gene of Vibrio cholerae encodes a growth-phase-dependent maltoporin. Mol Microbiol 10:891–901 10.1111/j.1365-2958.1993.tb00960.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. 2009. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology 155:498–512 10.1099/mic.0.022160-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1–32 10.1128/mmbr.49.1.1-32.1985. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forst S, Comeau D, Norioka S, Inouye M. 1987. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem 262:16433–16438. [PubMed] [Google Scholar]
- 19.Tanaka T, Saha SK, Tomomori C, Ishima R, Liu D, Tong KI, Park H, Dutta R, Qin L, Swindells MB, Yamazaki T, Ono AM, Kainosho M, Inouye M, Ikura M. 1998. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature 396:88–92 10.1038/23968. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, Qian H, Inouye M, Ikura M. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol 6:729–734 10.1038/11495. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Foo YH, Gao Y, Zhang H, Kenney LJ. 2015. Cytoplasmic sensing by the inner membrane histidine kinase EnvZ. Prog Biophys Mol Biol 118:119–129 10.1016/j.pbiomolbio.2015.04.005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appleman JA, Chen LL, Stewart V. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J Bacteriol 185:4872–4882 10.1128/JB.185.16.4872-4882.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H, Saha SK, Inouye M. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci USA 95:6728–6732 10.1073/pnas.95.12.6728. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty S, Mizusaki H, Kenney LJ. 2015. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13:e1002116 10.1371/journal.pbio.1002116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, Groisman EA. 2016. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol Microbiol 101:1024–1038 10.1111/mmi.13439. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eguchi Y, Utsumi R. 2014. Alkali metals in addition to acidic pH activate the EvgS histidine kinase sensor in Escherichia coli. J Bacteriol 196:3140–3149 10.1128/JB.01742-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen H, Aggarwal N, Ishionwu C, Hussain N, Parmar C, Jamshad M, Bavro VN, Lund PA. 2017. Structural and functional analysis of the Escherichia coli acid-sensing histidine kinase EvgS. J Bacteriol 199:199 10.1128/JB.00310-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. 1989. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245:1246–1249 10.1126/science.2476847. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Inouye M. 1991. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci USA 88:11057–11061 10.1073/pnas.88.24.11057. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casino P, Rubio V, Marina A. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139:325–336 10.1016/j.cell.2009.08.032. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Casino P, Miguel-Romero L, Marina A. 2014. Visualizing autophosphorylation in histidine kinases. Nat Commun 5:3258 10.1038/ncomms4258. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Ghosh M, Wang LC, Huber RG, Gao Y, Morgan LK, Tulsian NK, Bond PJ, Kenney LJ, Anand GS. 2019. Engineering an osmosensor by pivotal histidine positioning within disordered helices. Structure 27:P302–P314.E4 10.1016/j.str.2018.10.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940 10.1016/j.cell.2006.06.058. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Kishii R, Falzon L, Yoshida T, Kobayashi H, Inouye M. 2007. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J Biol Chem 282:26401–26408 10.1074/jbc.M701342200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Leonardo MR, Forst S. 1996. Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signals in Escherichia coli. Mol Microbiol 22:405–413 10.1046/j.1365-2958.1996.1271487.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Igo MM, Silhavy TJ. 1988. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol 170:5971–5973 10.1128/jb.170.12.5971-5973.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenney LJ. 1997. Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch Biochem Biophys 346:303–311 10.1006/abbi.1997.0315. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53:121–147. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foo YH, Spahn C, Zhang H, Heilemann M, Kenney LJ. 2015. Single cell super-resolution imaging of E. coli OmpR during environmental stress. Integr Biol 7:1297–1308 10.1039/c5ib00077g. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlösser A, Meldorf M, Stumpe S, Bakker EP, Epstein W. 1995. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J Bacteriol 177:1908–1910 10.1128/jb.177.7.1908-1910.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epstein W, Schultz SG. 1965. Cation transport in Escherichia coli. V. Regulation of cation content. J Gen Physiol 49:221–234 10.1085/jgp.49.2.221. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graeme-Cook KA. 1991. The regulation of porin expression in Escherichia coli: effect of turgor stress. FEMS Microbiol Lett 63:219–223 10.1111/j.1574-6968.1991.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 43.Cayley DS, Guttman HJ, Record MT Jr. 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys J 78:1748–1764 10.1016/S0006-3495(00)76726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auton M, Bolen DW. 2004. Additive transfer free energies of the peptide backbone unit that are independent of the model compound and the choice of concentration scale. Biochemistry 43:1329–1342 10.1021/bi035908r. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Auton M, Bolen DW. 2005. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA 102:15065–15068 10.1073/pnas.0507053102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Street TO, Bolen DW, Rose GD. 2006. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci USA 103:13997–14002 10.1073/pnas.0606236103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granett S, Villarejo M. 1982. Regulation of gene expression in Escherichia coli by the local anesthetic procaine. J Mol Biol 160:363–367 10.1016/0022-2836(82)90181-4. [DOI] [PubMed] [Google Scholar]
- 48.Rampersaud A, Inouye M. 1991. Procaine, a local anesthetic, signals through the EnvZ receptor to change the DNA binding affinity of the transcriptional activator protein OmpR. J Bacteriol 173:6882–6888 10.1128/jb.173.21.6882-6888.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Granett S, Villarejo M. 1981. Selective inhibition of carbohydrate transport by the local anesthetic procaine in Escherichia coli. J Bacteriol 147:289–296 10.1128/jb.147.2.289-296.1981. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Batchelor E, Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci USA 100:691–696 10.1073/pnas.0234782100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts DL, Bennett DW, Forst SA. 1994. Identification of the site of phosphorylation on the osmosensor, EnvZ, of Escherichia coli. J Biol Chem 269:8728–8733. [PubMed] [Google Scholar]
- 52.Delgado J, Forst S, Harlocker S, Inouye M. 1993. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol 10:1037–1047 10.1111/j.1365-2958.1993.tb00974.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Sullivan WT. 1994. The Salvation of Doug. GENErations 1(3). [Google Scholar]
- 54.Kellogg DR. 1994. The Demise of Bill. GENErations 2(1). [Google Scholar]
- 55.Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol 179:3729–3735 10.1128/jb.179.11.3729-3735.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo FD, Silhavy TJ. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol 222:567–580 10.1016/0022-2836(91)90497-T. [DOI] [PubMed] [Google Scholar]
- 57.Pioszak AA, Ninfa AJ. 2003. Mechanism of the PII-activated phosphatase activity of Escherichia coli NRII (NtrB): how the different domains of NRII collaborate to act as a phosphatase. Biochemistry 42:8885–8899 10.1021/bi030065p. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Pioszak AA, Ninfa AJ. 2003. Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein. J Bacteriol 185:1299–1315 10.1128/JB.185.4.1299-1315.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Wulf P, Lin EC. 2000. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J Bacteriol 182:1423–1426 10.1128/JB.182.5.1423-1426.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boesch KC, Silversmith RE, Bourret RB. 2000. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J Bacteriol 182:3544–3552 10.1128/JB.182.12.3544-3552.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9:570–575 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 62.Lima BP, Thanh Huyen TT, Bäsell K, Becher D, Antelmann H, Wolfe AJ. 2012. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J Biol Chem 287:32147–32160 10.1074/jbc.M112.365502. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. 2009. Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol 390:380–393 10.1016/j.jmb.2009.05.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. 2008. Rewiring the specificity of two-component signal transduction systems. Cell 133:1043–1054 10.1016/j.cell.2008.04.040. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aiba H, Nakasai F, Mizushima S, Mizuno T. 1989. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem 264:14090–14094. [PubMed] [Google Scholar]
- 66.Mattison K, Kenney LJ. 2002. Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem 277:11143–11148 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- 67.Kenney LJ. 2010. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol 13:168–176 10.1016/j.mib.2010.01.013. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kenney LJ, Bauer MD, Silhavy TJ. 1995. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci USA 92:8866–8870 10.1073/pnas.92.19.8866. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA 89:718–722 10.1073/pnas.89.2.718. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wanner BL. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol 174:2053–2058 10.1128/jb.174.7.2053-2058.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50 10.1128/MMBR.69.1.12-50.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batchelor E, Walthers D, Kenney LJ, Goulian M. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol 187:5723–5731 10.1128/JB.187.16.5723-5731.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189:5574–5581 10.1128/JB.00564-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733 10.1128/jb.179.24.7724-7733.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cosma CL, Danese PN, Carlson JH, Silhavy TJ, Snyder WB. 1995. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol 18:491–505 10.1111/j.1365-2958.1995.mmi_18030491.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398 10.1101/gad.9.4.387. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Møller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Conway T, Cohen PS. 2014. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun 82:670–682 10.1128/IAI.01149-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Möndel M, Schroeder BO, Zimmermann K, Huber H, Nuding S, Beisner J, Fellermann K, Stange EF, Wehkamp J. 2009. Probiotic E. coli treatment mediates antimicrobial human beta-defensin synthesis and fecal excretion in humans. Mucosal Immunol 2:166–172 10.1038/mi.2008.77. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 10.1126/science.277.5331.1453. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Peters JE, Thate TE, Craig NL. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J Bacteriol 185:2017–2021 10.1128/JB.185.6.2017-2021.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kitko RD, Wilks JC, Garduque GM, Slonczewski JL. 2010. Osmolytes contribute to pH homeostasis of Escherichia coli. PLoS One 5:e10078 10.1371/journal.pone.0010078. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Puttick J, Baker EN, Delbaere LT. 2008. Histidine phosphorylation in biological systems. Biochim Biophys Acta 1784:100–105 10.1016/j.bbapap.2007.07.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Jin T, Inouye M. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J Mol Biol 232:484–492 10.1006/jmbi.1993.1404. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Biemann HP, Koshland DE Jr. 1994. Aspartate receptors of Escherichia coli and Salmonella Typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry 33:629–634 10.1021/bi00169a002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Jung K, Hamann K, Revermann A. 2001. K+ stimulates specifically the autokinase activity of purified and reconstituted EnvZ of Escherichia coli. J Biol Chem 276:40896–40902 10.1074/jbc.M107871200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Gerken H, Charlson ES, Cicirelli EM, Kenney LJ, Misra R. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol Microbiol 72:1408–1422 10.1111/j.1365-2958.2009.06728.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerken H, Misra R. 2010. MzrA-EnvZ interactions in the periplasm influence the EnvZ/OmpR two-component regulon. J Bacteriol 192:6271–6278 10.1128/JB.00855-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506 10.1111/j.1365-2958.2008.06426.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liew A, Foo YH, Gao Y, Zangoui P, Singh MK, Gulvady R, Kenney LJ. 2019. Single cell, super-resolution imaging reveals an acid pH-dependent conformational switch in SsrB regulates SPI-2. eLife 8:e45311 10.7554/eLife.45311. [PubMed] [DOI] [PMC free article] [PubMed]
- 90.Spahn C, Glaesmann M, Gao Y, Foo YH, Lampe M, Kenney LJ, Heilemann M. 2017. Sequential super-resolution imaging of bacterial regulatory proteins, the nucleoid and the cell membrane in single, fixed E. coli cells. In Espeli O (ed), The Bacterial Nucleoid. Methods in Molecular Biology, vol 1624. Humana Press, New York, NY. 10.1007/978-1-4939-7098-8_20 [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Ghosh M, Wang LC, Ramesh R, Morgan LK, Kenney LJ, Anand GS. 2017. Lipid-mediated regulation of embedded receptor kinases via parallel allosteric relays. Biophys J 112:643–654 10.1016/j.bpj.2016.12.027. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kato M, Aiba H, Tate S, Nishimura Y, Mizuno T. 1989. Location of phosphorylation site and DNA-binding site of a positive regulator, OmpR, involved in activation of the osmoregulatory genes of Escherichia coli. FEBS Lett 249:168–172 10.1016/0014-5793(89)80617-9. [DOI] [PubMed] [Google Scholar]
- 93.Kenney LJ. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5:135–141 10.1016/S1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 94.Martínez-Hackert E, Stock AM. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109–124 10.1016/S0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 95.Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. 2008. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem 283:8664–8677 10.1074/jbc.M705550200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mizuno T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4:161–168 10.1093/dnares/4.2.161. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Bourret RB. 2010. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 13:142–149 10.1016/j.mib.2010.01.015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mattison K, Oropeza R, Byers N, Kenney LJ. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol 315:497–511 10.1006/jmbi.2001.5222. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Huang KJ, Igo MM. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol 262:615–628 10.1006/jmbi.1996.0540. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Huang KJ, Schieberl JL, Igo MM. 1994. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J Bacteriol 176:1309–1315 10.1128/jb.176.5.1309-1315.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kondo H, Miyaji T, Suzuki M, Tate S, Mizuno T, Nishimura Y, Tanaka I. 1994. Crystallization and X-ray studies of the DNA-binding domain of OmpR protein, a positive regulator involved in activation of osmoregulatory genes in Escherichia coli. J Mol Biol 235:780–782 10.1006/jmbi.1994.1032. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Martínez-Hackert E, Stock AM. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol 269:301–312 10.1006/jmbi.1997.1065. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Kim SK, Makino K, Amemura M, Nakata A, Shinagawa H. 1995. Mutational analysis of the role of the first helix of region 4.2 of the sigma 70 subunit of Escherichia coli RNA polymerase in transcriptional activation by activator protein PhoB. Mol Gen Genet 248:1–8 10.1007/BF02456607. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Makino K, Amemura M, Kim SK, Nakata A, Shinagawa H. 1993. Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev 7:149–160 10.1101/gad.7.1.149. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Pratt LA, Silhavy TJ. 1994. OmpR mutants specifically defective for transcriptional activation. J Mol Biol 243:579–594 10.1016/0022-2836(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 106.Head CG, Tardy A, Kenney LJ. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol 281:857–870 10.1006/jmbi.1998.1985. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Brosse A, Korobeinikova A, Gottesman S, Guillier M. 2016. Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res 44:9650–9666 10.1093/nar/gkw642. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carroll RK, Liao X, Morgan LK, Cicirelli EM, Li Y, Sheng W, Feng X, Kenney LJ. 2009. Structural and functional analysis of the C-terminal DNA binding domain of the Salmonella Typhimurium SPI-2 response regulator SsrB. J Biol Chem 284:12008–12019 10.1074/jbc.M806261200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stincone A, Daudi N, Rahman AS, Antczak P, Henderson I, Cole J, Johnson MD, Lund P, Falciani F. 2011. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res 39:7512–7528 10.1093/nar/gkr338. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng X, Oropeza R, Kenney LJ. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol 48:1131–1143 10.1046/j.1365-2958.2003.03502.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Lee AK, Detweiler CS, Falkow S. 2000. OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. J Bacteriol 182:771–781 10.1128/JB.182.3.771-781.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bachhawat P, Swapna GV, Montelione GT, Stock AM. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13:1353–1363 10.1016/j.str.2005.06.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blanco AG, Sola M, Gomis-Rüth FX, Coll M. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701–713 10.1016/S0969-2126(02)00761-X. [DOI] [PubMed] [Google Scholar]
- 114.Maris AE, Walthers D, Mattison K, Byers N, Kenney LJ. 2005. The response regulator OmpR oligomerizes via β-sheets to form head-to-head dimers. J Mol Biol 350:843–856 10.1016/j.jmb.2005.05.057. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Brissette RE, Tsung K, Inouye M. 1992. Mutations in a central highly conserved non-DNA-binding region of OmpR, an Escherichia coli transcriptional activator, influence its DNA-binding ability. J Bacteriol 174:4907–4912 10.1128/jb.174.15.4907-4912.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nakashima K, Kanamaru K, Aiba H, Mizuno T. 1991. Signal transduction and osmoregulation in Escherichia coli. A novel type of mutation in the phosphorylation domain of the activator protein, OmpR, results in a defect in its phosphorylation-dependent DNA binding. J Biol Chem 266:10775–10780. [PubMed] [Google Scholar]
- 117.Ames SK, Frankema N, Kenney LJ. 1999. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci USA 96:11792–11797 10.1073/pnas.96.21.11792. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qin L, Yoshida T, Inouye M. 2001. The critical role of DNA in the equilibrium between OmpR and phosphorylated OmpR mediated by EnvZ in Escherichia coli. Proc Natl Acad Sci USA 98:908–913 10.1073/pnas.98.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li J, Swanson RV, Simon MI, Weis RM. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34:14626–14636 10.1021/bi00045a003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Harlocker SL, Bergstrom L, Inouye M. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem 270:26849–26856 10.1074/jbc.270.45.26849. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Maeda S, Mizuno T. 1990. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J Bacteriol 172:501–503 10.1128/jb.172.1.501-503.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rampersaud A, Harlocker SL, Inouye M. 1994. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem 269:12559–12566. [PubMed] [Google Scholar]
- 123.Ostrow KS, Silhavy TJ, Garrett S. 1986. cis-acting sites required for osmoregulation of ompF expression in Escherichia coli K-12. J Bacteriol 168:1165–1171 10.1128/jb.168.3.1165-1171.1986. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maeda S, Takayanagi K, Nishimura Y, Maruyama T, Sato K, Mizuno T. 1991. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: a study involving synthetic OmpR-binding sequences. J Biochem 110:324–327 10.1093/oxfordjournals.jbchem.a123579. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Harrison-McMonagle P, Denissova N, Martínez-Hackert E, Ebright RH, Stock AM. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J Mol Biol 285:555–566 10.1006/jmbi.1998.2375. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Barbieri CM, Wu T, Stock AM. 2013. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J Mol Biol 425:1612–1626 10.1016/j.jmb.2013.02.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ozawa Y, Mizuno T, Mizushima S. 1987. Roles of the Pribnow box in positive regulation of the ompC and ompF genes in Escherichia coli. J Bacteriol 169:1331–1334 10.1128/jb.169.3.1331-1334.1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Igarashi K, Hanamura A, Makino K, Aiba H, Aiba H, Mizuno T, Nakata A, Ishihama A. 1991. Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc Natl Acad Sci USA 88:8958–8962 10.1073/pnas.88.20.8958. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sharif TR, Igo MM. 1993. Mutations in the alpha subunit of RNA polymerase that affect the regulation of porin gene transcription in Escherichia coli K-12. J Bacteriol 175:5460–5468 10.1128/jb.175.17.5460-5468.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Slauch JM, Russo FD, Silhavy TJ. 1991. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the alpha subunit of RNA polymerase. J Bacteriol 173:7501–7510 10.1128/jb.173.23.7501-7510.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol 37:687–695 10.1046/j.1365-2958.2000.01972.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. 2002. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297:1562–1566 10.1126/science.1076376. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Boucher PE, Maris AE, Yang MS, Stibitz S. 2003. The response regulator BvgA and RNA polymerase alpha subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol Cell 11:163–173 10.1016/S1097-2765(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 134.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213 10.1006/jmbi.1999.3161. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Chen H, Tang H, Ebright RH. 2003. Functional interaction between RNA polymerase alpha subunit C-terminal domain and sigma70 in UP-element- and activator-dependent transcription. Mol Cell 11:1621–1633 10.1016/S1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 136.Czarniecki D, Noel RJ Jr, Reznikoff WS. 1997. The –45 region of the Escherichia colilac promoter: CAP-dependent and CAP-independent transcription. J Bacteriol 179:423–429 10.1128/jb.179.2.423-429.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Flatow U, Rajendrakumar GV, Garges S. 1996. Analysis of the spacer DNA between the cyclic AMP receptor protein binding site and the lac promoter. J Bacteriol 178:2436–2439 10.1128/jb.178.8.2436-2439.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. 2003. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the alpha C-terminal domain and sigma region 4. Genes Dev 17:1293–1307 10.1101/gad.1079403. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kato N, Aiba H, Mizuno T. 1996. Suppressor mutations in alpha-subunit of RNA polymerase for a mutant of the positive regulator, OmpR, in Escherichia coli. FEMS Microbiol Lett 139:175–180. [DOI] [PubMed] [Google Scholar]
- 140.Bowrin V, Brissette R, Tsung K, Inouye M. 1994. The alpha subunit of RNA polymerase specifically inhibits expression of the porin genes ompF and ompC in vivo and in vitro in Escherichia coli. FEMS Microbiol Lett 115:1–6. [DOI] [PubMed] [Google Scholar]
- 141.Aiba H, Kato N, Tsuzuki M, Mizuno T. 1994. Mechanism of gene activation by the Escherichia coli positive regulator, OmpR: a mutant defective in transcriptional activation. FEBS Lett 351:303–307 10.1016/0014-5793(94)00846-9. [DOI] [PubMed] [Google Scholar]
- 142.Kato N, Tsuzuki M, Aiba H, Mizuno T. 1995. Gene activation by the Escherichia coli positive regulator OmpR: a mutational study of the DNA-binding domain of OmpR. Mol Gen Genet 248:399–406 10.1007/BF02191639. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Itou H, Tanaka I. 2001. The OmpR-family of proteins: insight into the tertiary structure and functions of two-component regulator proteins. J Biochem 129:343–350 10.1093/oxfordjournals.jbchem.a002863. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Nogami T, Mizuno T, Mizushima S. 1985. Construction of a series of ompF-ompC chimeric genes by in vivo homologous recombination in Escherichia coli and characterization of the translational products. J Bacteriol 164:797–801. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7:1505–1515 10.1016/S0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- 146.Cho HS, Lee SY, Yan D, Pan X, Parkinson JS, Kustu S, Wemmer DE, Pelton JG. 2000. NMR structure of activated CheY. J Mol Biol 297:543–551 10.1006/jmbi.2000.3595. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Halkides CJ, McEvoy MM, Casper E, Matsumura P, Volz K, Dahlquist FW. 2000. The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry 39:5280–5286 10.1021/bi9925524. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Lewis RJ, Brannigan JA, Muchová K, Barák I, Wilkinson AJ. 1999. Phosphorylated aspartate in the structure of a response regulator protein. J Mol Biol 294:9–15 10.1006/jmbi.1999.3261. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Huang L, Tsui P, Freundlich M. 1990. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J Bacteriol 172:5293–5298 10.1128/jb.172.9.5293-5298.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sheridan SD, Benham CJ, Hatfield GW. 1998. Activation of gene expression by a novel DNA structural transmission mechanism that requires supercoiling-induced DNA duplex destabilization in an upstream activating sequence. J Biol Chem 273:21298–21308 10.1074/jbc.273.33.21298. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Tran VK, Oropeza R, Kenney LJ. 2000. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J Mol Biol 299:1257–1270 10.1006/jmbi.2000.3809. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Tsui P, Huang L, Freundlich M. 1991. Integration host factor binds specifically to multiple sites in the ompB promoter of Escherichia coli and inhibits transcription. J Bacteriol 173:5800–5807 10.1128/jb.173.18.5800-5807.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291 10.1046/j.1365-2958.2002.03170.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972 10.1128/JB.185.16.4956-4972.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hirakawa H, Nishino K, Hirata T, Yamaguchi A. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Bacteriol 185:1851–1856 10.1128/JB.185.6.1851-1856.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shin S, Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177:4696–4702 10.1128/jb.177.16.4696-4702.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gerstel U, Park C, Römling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol 49:639–654 10.1046/j.1365-2958.2003.03594.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol 183:7213–7223 10.1128/JB.183.24.7213-7223.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol 180:2442–2449. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fernández-Mora M, Oropeza R, Puente JL, Calva E. 1995. Isolation and characterization of ompS1, a novel Salmonella Typhi outer membrane protein-encoding gene. Gene 158:67–72 10.1016/0378-1119(95)00171-2. [DOI] [PubMed] [Google Scholar]
- 161.Oropeza R, Sampieri CL, Puente JL, Calva E. 1999. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella Typhi: a novel regulatory mechanism that involves OmpR. Mol Microbiol 32:243–252 10.1046/j.1365-2958.1999.01329.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Bang IS, Audia JP, Park YK, Foster JW. 2002. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol Microbiol 44:1235–1250 10.1046/j.1365-2958.2002.02937.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 163.Bang IS, Kim BH, Foster JW, Park YK. 2000. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol 182:2245–2252 10.1128/JB.182.8.2245-2252.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bernardini ML, Fontaine A, Sansonetti PJ. 1990. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J Bacteriol 172:6274–6281 10.1128/jb.172.11.6274-6281.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dorman CJ, Chatfield S, Higgins CF, Hayward C, Dougan G. 1989. Characterization of porin and ompR mutants of a virulent strain of Salmonella Typhimurium: ompR mutants are attenuated in vivo. Infect Immun 57:2136–2140. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dorrell N, Li SR, Everest PH, Dougan G, Wren BW. 1998. Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol Lett 165:145–151 10.1111/j.1574-6968.1998.tb13139.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 167.Lindgren SW, Stojiljkovic I, Heffron F. 1996. Macrophage killing is an essential virulence mechanism of Salmonella Typhimurium. Proc Natl Acad Sci USA 93:4197–4201 10.1073/pnas.93.9.4197. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tucker DL, Tucker N, Conway T. 2002. Gene expression profiling of the pH response in Escherichia coli. J Bacteriol 184:6551–6558 10.1128/JB.184.23.6551-6558.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Guillier M, Gottesman S. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59:231–247 10.1111/j.1365-2958.2005.04929.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Quinn HJ, Cameron AD, Dorman CJ. 2014. Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet 10:e1004215 10.1371/journal.pgen.1004215. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cheeseman GC, Fuller R. 1968. Changes in the pH activity profile of the lysine decarboxylase during incubation of Escherichia coli. J Appl Bacteriol 31:253–258 10.1111/j.1365-2672.1968.tb00365.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 172.Vivijs B, Aertsen A, Michiels CW. 2016. Identification of genes required for growth of Escherichia coli MG1655 at moderately low pH. Front Microbiol 7:1672 10.3389/fmicb.2016.01672. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Blomfield IC, McClain MS, Princ JA, Calie PJ, Eisenstein BI. 1991. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol 173:5298–5307 10.1128/jb.173.17.5298-5307.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Miller J. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 175.Armitage JP. 1999. Bacterial tactic responses. Adv Microb Physiol 41:229–289 10.1016/S0065-2911(08)60168-X. [DOI] [PubMed] [Google Scholar]
- 176.Falke JJ, Hazelbauer GL. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265 10.1016/S0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Prüss BM. 1998. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch Microbiol 170:141–146 10.1007/s002030050626. [PubMed] [DOI] [PubMed] [Google Scholar]
- 178.Prüss BM, Wolfe AJ. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol 12:973–984 10.1111/j.1365-2958.1994.tb01085.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Wolfe AJ, Chang DE, Walker JD, Seitz-Partridge JE, Vidaurri MD, Lange CF, Prüss BM, Henk MC, Larkin JC, Conway T. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol Microbiol 48:977–988 10.1046/j.1365-2958.2003.03457.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209 10.1146/annurev.micro.56.012302.160705. [PubMed] [DOI] [PubMed] [Google Scholar]
- 181.Desai SK, Kenney LJ. 2017. To ∼P or not to ∼P? Non-canonical activation by two-component response regulators. Mol Microbiol 103:203–213 10.1111/mmi.13532. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Desai SK, Padmanabhan A, Harshe S, Zaidel-Bar R, Kenney LJ. 2019. Salmonella biofilms program innate immunity for persistence in C. elegans. Proc Natl Acad Sci USA 116:12462–12467 10.1073/pnas.1822018116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Desai SK, Winardhi RS, Periasamy S, Dykas MM, Jie Y, Kenney LJ. 2016. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. eLife 5:e10747 10.7554/eLife.10747. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Feng X, Walthers D, Oropeza R, Kenney LJ. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54:823–835 10.1111/j.1365-2958.2004.04317.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, Keane T, Maskell D, Hinton JC, Dougan G, Kingsley RA. 2013. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol Microbiol 87:526–538 10.1111/mmi.12111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shimada T, Takada H, Yamamoto K, Ishihama A. 2015. Expanded roles of two-component response regulator OmpR in Escherichia coli: genomic SELEX search for novel regulation targets. Genes Cells 20:915–931 10.1111/gtc.12282. [PubMed] [DOI] [PubMed] [Google Scholar]
- 187.Cameron AD, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8:e1002615 10.1371/journal.pgen.1002615. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Aiba H, Mizuno T, Mizushima S. 1989. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem 264:8563–8567. [PubMed] [Google Scholar]
- 189.Huyghues-Despointes BM, Baldwin RL. 1997. Ion-pair and charged hydrogen-bond interactions between histidine and aspartate in a peptide helix. Biochemistry 36:1965–1970 10.1021/bi962546x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Ferris HU, Coles M, Lupas AN, Hartmann MD. 2014. Crystallographic snapshot of the Escherichia coli EnvZ histidine kinase in an active conformation. J Struct Biol 186:376–379 10.1016/j.jsb.2014.03.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 191.Hall MN, Silhavy TJ. 1981. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol 151:1–15 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- 192.Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. 2010. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet 6:e1001220 10.1371/journal.pgen.1001220. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Conley MP, Berg HC, Tawa P, Stewart RC, Ellefson DD, Wolfe AJ. 1994. pH dependence of CheA autophosphorylation in Escherichia coli. J Bacteriol 176:3870–3877 10.1128/jb.176.13.3870-3877.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Batchelor E, Goulian M. 2006. Imaging OmpR localization in Escherichia coli. Mol Microbiol 59:1767–1778 10.1111/j.1365-2958.2006.05048.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 195.Heilemann M. 2010. Fluorescence microscopy beyond the diffraction limit. J Biotechnol 149:243–251 10.1016/j.jbiotec.2010.03.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 196.Gao Y, Spahn C, Heilemann M, Kenney LJ. 2018. The pearling transition provides evidence of force-driven endosomal tubulation during Salmonella infection. MBio 9:e01083-18 10.1128/mBio.01083-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gao Y, Foo YH, Winardhi RS, Tang Q, Yan J, Kenney LJ. 2017. A novel DNA binding mode of H-NS drives gene silencing in single cells. Proc Natl Acad Sci USA 10.1073/pnas.1716721114. [PubMed] [DOI] [PMC free article] [PubMed]
- 198.Kamiyama D, Sekine S, Barsi-Rhyne B, Hu J, Chen B, Gilbert LA, Ishikawa H, Leonetti MD, Marshall WF, Weissman JS, Huang B. 2016. Versatile protein tagging in cells with split fluorescent protein. Nat Commun 7:11046 10.1038/ncomms11046. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Libby EA, Ekici S, Goulian M. 2010. Imaging OmpR binding to native chromosomal loci in Escherichia coli. J Bacteriol 192:4045–4053 10.1128/JB.00344-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


