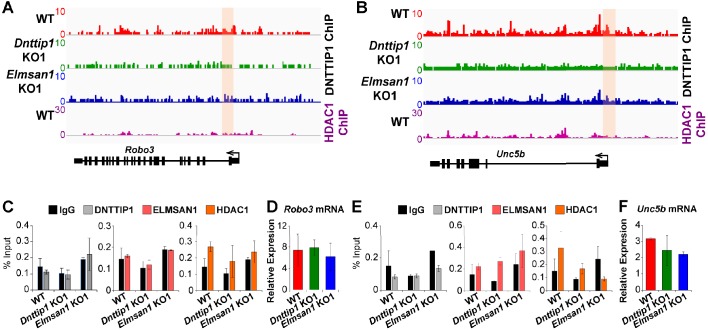
(
A, B) ChIP-seq profiles of the (
A)
Robo3 and (
B)
Unc5b loci for DNTTIP1 in WT,
Dnttip1 KO1 and
Elmsan1 KO1 mESCs and for HDAC1 in WT mESCs. Promoter regions used for manual ChIP experiments in (
C) and (
E) are highlighted by orange boxes. DNTTIP1 ChIP-seq peaks were determined based on the average of two replicates each from WT mESCs and based on the average of two replicates each from the
Dnttip1 KO1 and
Elmsan1 KO1 clone, respectively. HDAC1 ChIP-seq data were obtained from
GSE55437. (
C) qPCR from manual ChIP experiments against DNTTIP1, ELMSAN1 and HDAC1 from WT,
Dnttip1 KO1 and
Elmsan1 KO1 NE targeting the promoter of the
Robo3 locus as highlighted in (
A). IgG was used as a control antibody. (
D) qRT-PCR for
Robo3 mRNA in WT,
Dnttip1 KO1 and
Elmsan1 KO1 NE after 12 days of differentiation. Expression was normalized to Gapdh. (
E) qPCR from manual ChIP experiments against DNTTIP1, ELMSAN1 and HDAC1 from WT,
Dnttip1 KO1 and
Elmsan1 KO1 NE targeting the promoter of the
Unc5b locus as highlighted in (
B). IgG was used as a control antibody. (
F) qRT-PCR for
Unc5b mRNA in WT,
Dnttip1 KO1 and
Elmsan1 KO1 NE after 12 days of differentiation. Expression was normalized to
Gapdh.