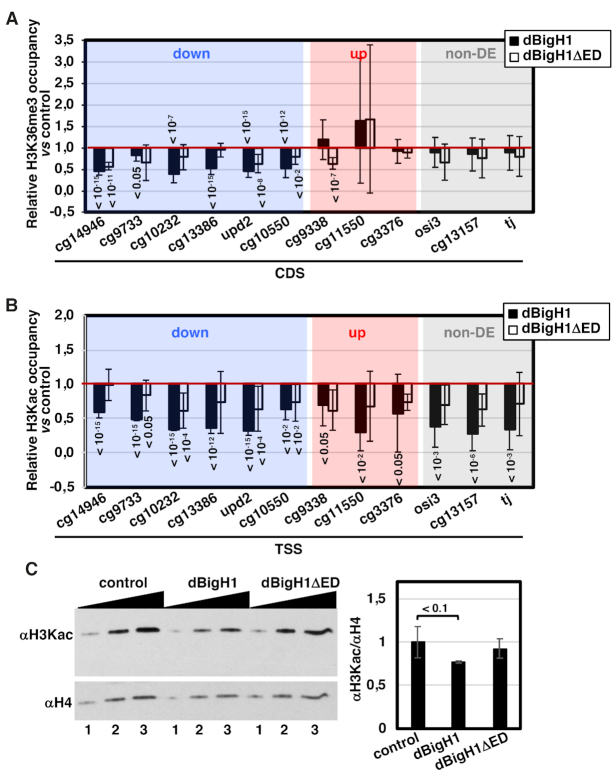
Figure 7.
dBigH1 perturbs the pattern of histone modifications. (A) The relative H3K36me3 occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at CDS of the indicated genes in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 3; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (B) The relative H3Kac occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at TSS of the indicated genes in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 2; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (C) In the left, WB analysis with αH3Kac antibodies of crosslinked chromatin prepared from control mock-induced cells and cells expressing full-length dBigH1 and the truncated dBigH1ΔED forms. Increasing amounts of chromatin are analyzed (lanes 1–3). αH4 antibodies were used for normalization. Quantitative analysis of the results is shown in the right (N = 3; error bars are SD; two tailed t-test, P-values are indicated).