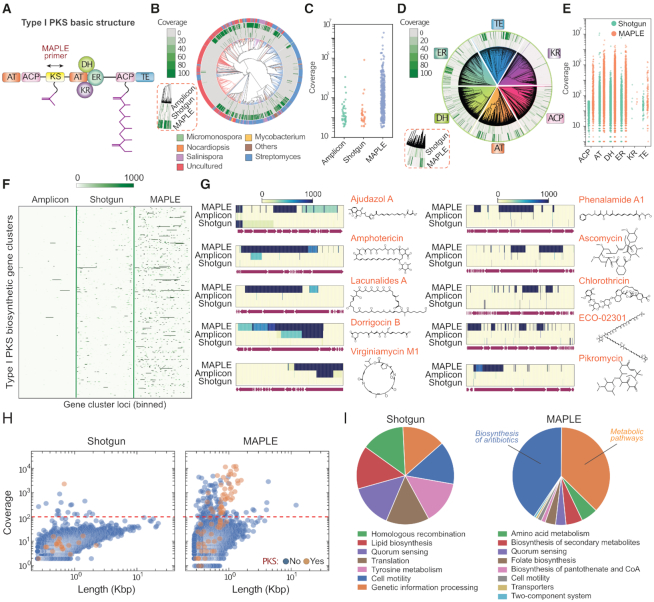
Figure 3.
MAPLE enriches PKS gene clusters and associated pathways from an Antarctic soil metagenomic library. (A) Basic modular structure of type I PKS gene cluster. The arrow indicates MAPLE primers targeting the KS gene. AT: acyltransferase; ACP: acyl carrier protein; KS: keto-synthase; DH: dehydratase; ER: enoyl-reductase; KR: keto-reductase; TE: thioesterase. (B) Reads mapping to the phylogenetic tree of KS genes from the NCBI database. Methods are shown in the red dashed box. Branch colors and outermost ring indicate genus from which a KS domain's gene cluster originates, with labels at the bottom. (C) Identification and coverage of KS genes. Each dot represents an identified KS gene. (D) Reads mapping to phylogenetic trees of the six other genes in PKS gene clusters. Methods are shown in the red dashed box. (E) Identification and coverage of six other genes in PKS gene clusters. Each dot represents an identified gene. (F) Heatmap illustrating coverage of selected PKS gene clusters. The x-axis is normalized positions of every gene cluster, with intensity indicating sequencing depth at that position. (G) Heatmaps for ten representative gene clusters for the synthesis of antibiotics with corresponding chemical structures on the right. (H) Scatter plots illustrating the distribution of de novo assembled contigs versus length and reads coverage. Contigs with and without PKS annotations are differentiated by color. The red dashed line indicates the threshold to separate on- and off-target contigs in MAPLE. (I) Pie charts comparing the distribution of the pathways identified by the two methods.