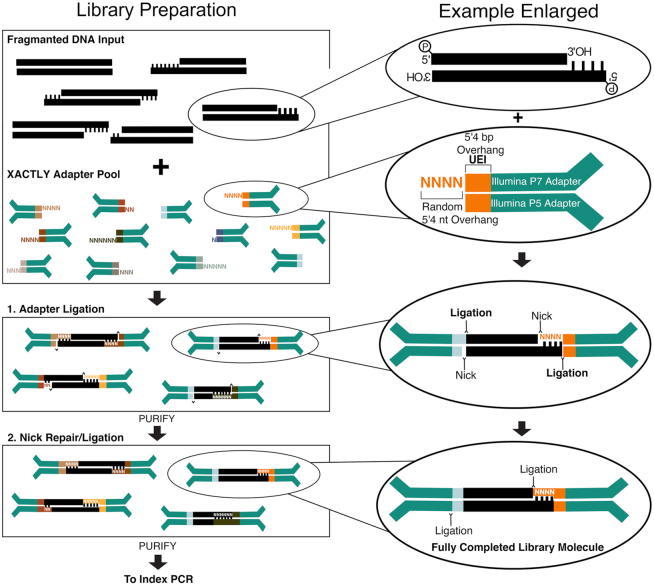
Figure 1.
Schematic overview of the XACTLY method designed for use on Illumina sequencing platforms. Modified and dephosphorylated Illumina Y-adapters contain a 7nt-barcode – called a Unique End Identifier (short multi-colored rectangles) – that denotes a discrete terminus type and length. The length of the overhang is represented by the corresponding number of random bases (shown as Ns), with the exception of a blunt end (no additional bases). Template DNA is first phosphorylated in preparation for adapter ligation but not end-polished, leaving intact any natively sticky or blunt ends. The XACTLY adapter set is then hybridized and ligated to the template DNA. A second round of phosphorylation and ligation seals the nicks present due to the 5′ dephosphorylated adapters. Libraries are then ready for PCR amplification using Illumina-compatible indexing primers (P5 and P7).