Figure 4.
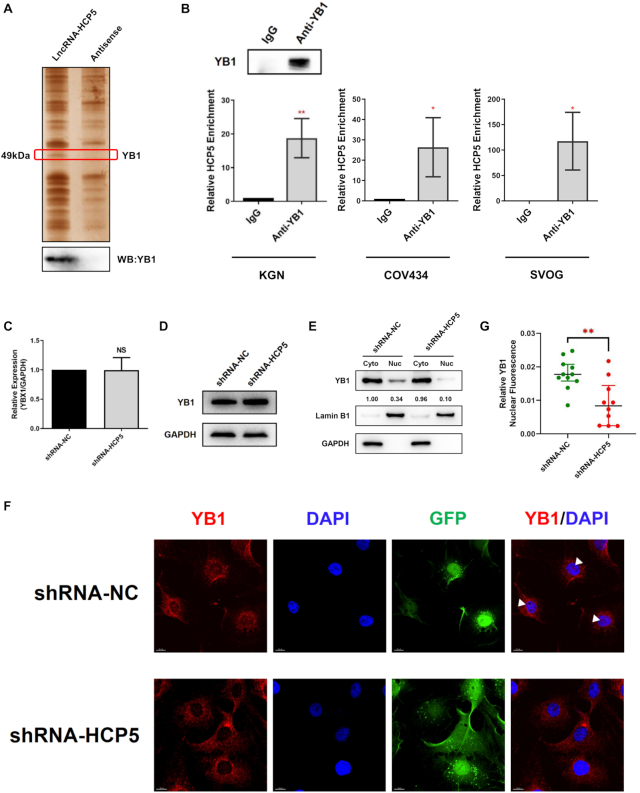
HCP5 directly binds to YB1 and is essential its nuclear localization. (A) Detection of HCP5-binding proteins by RNA pull-down assays. A specific band of ∼50 kDa (red box) was specific pulled down by HCP5 and subsequently identified as YB1 using mass spectrometry (Upper). Western blot showing the specific association between YB1 and HCP5 in the samples obtained from RNA pull-down (lower). The antisense transcript of HCP5 was used as a negative control. (B) Confirmation of the interaction between YB1 and HCP5 by RNA-binding protein immunoprecipitation (RIP) using YB1 antibody in KGN, COV434 and SVOG cells. Results are expressed as the mean ± SD (n = 3). *P < 0.05 and **P < 0.01. Two-tailed Student's t-test. (C) YB1 mRNA levels were analyzed by qRT-PCR after HCP5 silencing by shRNA in KGN cells. Values of qRT-PCR were obtained from triplicates and expressed as the mean ± SD (n = 3). Two-tailed Student's t-test. (D) YB1 protein levels were analyzed by western blot after HCP5 knockdown in KGN cells. Data shown represent three independent experiments. (E) Subcellular localization of YB1 protein was detected by western blot after HCP5 knockdown in KGN cells. Lamin B1 was used as nuclear control. GAPDH was used as cytoplasmic control. (F) Subcellular localization of YB1 protein was confirmed by immunofluorescence assay after HCP5 silencing. White triangles indicate representative YB1 expression in the nucleus. (G) Quantification of nuclear immunofluorescence intensity (mean gray value) in HCP5- knockdown (n = 10) and negative control (n = 11) KGN cells. Data are presented as the median ± interquartile range. **P < 0.01. Two-tailed Mann–Whitney U-test.