Figure 3.
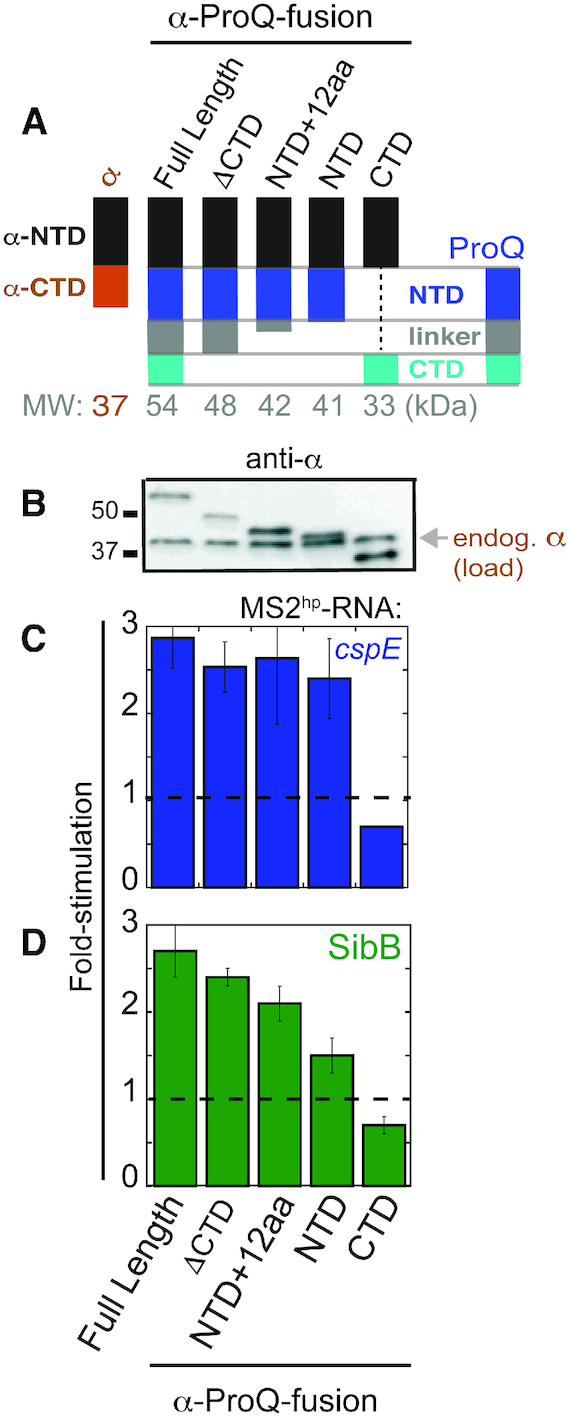
NTD is the primary site of interaction with cspE and SibB RNAs in vivo. (A) Schematic of α-ProQ domain-truncation mutants used in B3H assays, (B) Western blot with anti-RpoA antibody showing expression of α-ProQ truncations in lysates from samples in (C) and (D). The position of full-length endogenous RpoA (α; 37 kDa) and two molecular weight markers are indicated. Results of B3H assays detecting interactions between α-ProQ truncations and (C) cspE and (D) SibB RNAs. β-galactosidase assays were performed with Δhfq reporter strain cells containing three compatible plasmids: one that encoded λCI alone or the CI-MS2CP fusion protein, another that encoded α or an α-fusion protein (α-ProQFL (full-length; residues = 2–232), α-ProQΔCTD (residues = 2–176), α-ProQNTD+12aa (residues = 2–131), α-ProQNTD (residues = 2–119), or α-ProQCTD (residues = 181–232)), and a third that encoded a hybrid RNA (MS2hp-cspE or MS2hp-SibB) or an RNA that contained only the MS2hp moiety. The cells were grown in the presence of 0.2% arabinose and 50 μM IPTG (see Methods). Absolute β-gal values of a representative dataset are shown in Supplementary Figure S5B. Quantification of the Western blot is shown in Supplementary Figure S5C.
