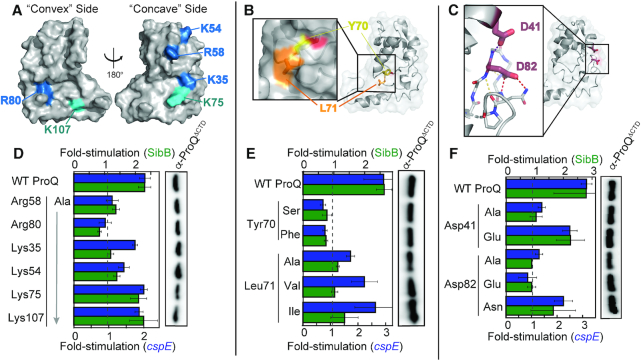
Figure 4.
Effects of specific disruptive amino acid substitutions on B3H interactions. Positions of (A) basic, (B) hydrophobic and (C) acidic residues targeted for site-directed mutagenesis shown on ProQ NTD Structure (PDB ID: 5nb9) (28). Residue coloring, used throughout: highly conserved basic, blue; less conserved basic, cyan; hydrophobic, orange; aromatic, yellow; acidic, red). (D–F, left) Inset in (B) shows surface representation with the hydroxyl group of Tyr70 colored red. Inset in (C) shows β-3,4 hairpin in stick representation, with atoms colored by element. Dashed lines indicate potential hydrogen bonds, involving the polypeptide backbone (yellow), Asp82 (red) or Asp41 (purple). Results of B3H assays showing effects on ProQ–RNA interactions of alanine mutations at (D) basic, (E) hydrophobic and (F) acidic residues. β-galactosidase assays were performed with Δhfq reporter strain cells containing three compatible plasmids: one that encoded λCI or the CI-MS2CP fusion protein, another that encoded α or an α-ProQΔCTD fusion protein (wild type, WT, or the indicated mutant), and a third that encoded a hybrid RNA (MS2hp-cspE or MS2hp-SibB) or an RNA that contained only the MS2hp moiety. (D, E) Cells were grown in the presence of 0.2% arabinose and 50 μM IPTG. (F) Cells were grown in the presence of 0.2% arabinose. The bar graph shows the fold-stimulation over basal levels as averages and standard deviations of values collected from two independent experiments conducted in triplicate across multiple days. Absolute β-gal values of a representative dataset are shown in Supplementary Figure S7. (D–F, right) Western blot to compare steady-state expression levels of mutant α-ProQΔCTD fusion proteins. Lysates were taken from the corresponding β-gal experiment containing MS2hp-cspE and all other hybrid components at 50 μM. Following electrophoresis and transfer, membranes were probed with anti-ProQ antibody (see Supplementary Figure S3). Loading controls and quantification of blots are shown in Supplementary Figure S8.