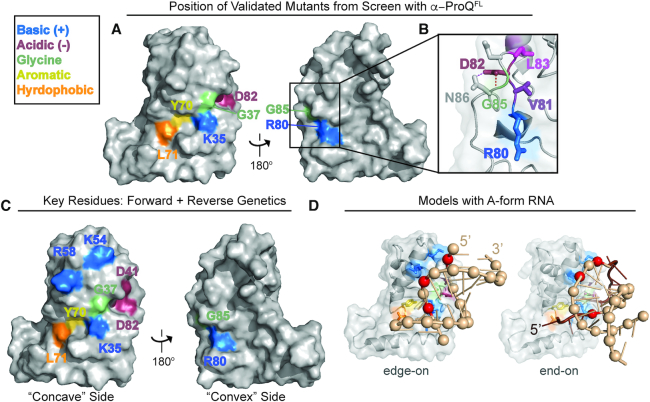
Figure 5.
Validated genetic-screen results and model for ProQ–dsRNA interactions. (A) Surface representation of ProQ NTD structure (PDB ID: 5nb9) (28), viewed from concave (left) or convex (right) surface, showing residues at which substitutions were found to disrupt RNA binding in B3H screens of mutagenized α-ProQFL plasmids and at which substitution with alanine has been confirmed to be sufficient to disrupt binding (Figure 4 and Supplementary Figure S10E; residue coloring: basic, blue; hydrophobic, orange; aromatic, yellow; acidic, red; polar: purple; glycine: green). (B) Inset shows close-up view of β3/4 hairpin, viewed from the convex face. Under a transparent surface representation, the polypeptide backbone is shown as a cartoon and amino-acid side chains are represented sticks, colored as in (A). Side chains for residues Asp84 and Asn86 (not identified as RNA-binding residues in screen) are shown as gray sticks. (C) Summary of results from both site-directed and unbiased mutagenesis experiments. Surface representation of ProQ NTD structure, viewed from concave (left) or convex (right) surface, showing all residues identified in this study as necessary for strong RNA interactions in vivo whether from site-directed mutagenesis (Figure 4) or a forward genetic screen (A) and colored as in (A). (D) Preliminary structural model for ProQ NTD recognition of dsRNA. Transparent surface and cartoon representation of ProQ NTD viewed from concave face, hand-docked in PyMol (version 1.6.2) and COOT (version 0.8.9.2) (52) to a 12-bp RNA duplex (PDB ID: 5DA6) (51) using only rigid rotations of the protein and RNA structure (left) or to a 10 bp RNA duplex and adjacent ssRNA from a RydC crystal structure (PDB ID: 4v2s) (50), using rigid rotations of the dsRNA, and rotations around phosphates of ssRNA (right). dsRNA is shown as a tan cartoon with phosphates as spheres, and adjacent ssRNA is shown as a brown cartoon. Three phosphates that have a suitable geometry to interact with basic residues (Lys35, Lys54 and Arg58) are colored in red. Two structural models that place these phosphates in an appropriate orientation to interact with these concave-face basic residues are shown: one in which an RNA duplex interacts with ProQ in an edge-on manner (left) and one in which it interacts in an end-on manner (right). Both of these models are consistent with our genetic data, though an end-on interaction is more consistent with biochemical data for FinO-domain proteins (see text). ProQ and residues are colored as in (C) with side chains of RNA-binding residues shown as sticks.