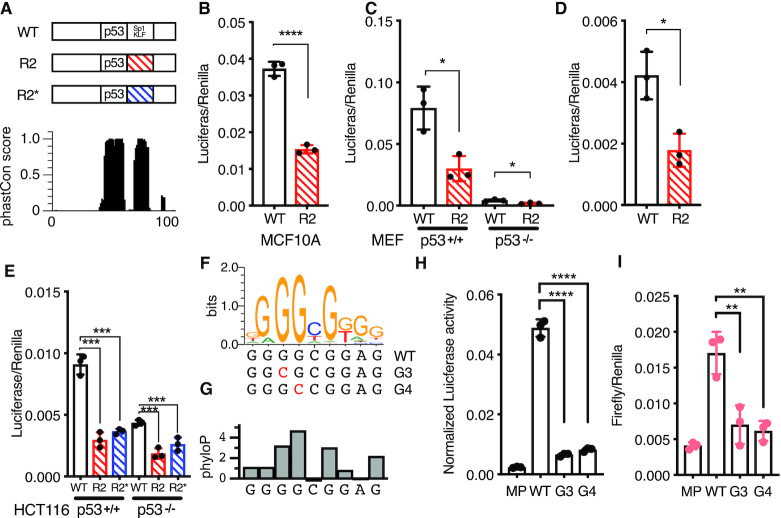
Figure 4.
(A) (top) Schematic of the 100 bp Region 36/CCNG1 enhancer. R2 position is annotated with the predicted motif ‘SP1/KLF’ 20 bp 3′ adjacent to the p53RE and (bottom) per-basepair vertebrate phastCon score. (B) Normalized Luciferase activity (test sequence Firefly versus constitutive Renilla) for WT Region36/CCNG and the R2 version in the MCF10A mammary epithelial cell line (****P < 0.0001 paired t-test). (C) Normalized Luciferase activity (test sequence Firefly versus constitutive Renilla) for WT Region36/CCNG and the R2 version in either p53+/+ or p53−/− mouse embryonic fibroblasts (*P < 0.05, paired t-test). (D) Rescaled view of p53−/− reporter assay data from (C) depicting normalized Luciferase data for the wild-type or R2 version of the CCNG1 enhancer in p53−/− mouse embryonic fibroblasts (*P < 0.05, paired t-test). (E) Normalized luciferase activity for either the wild-type, R2 or R2* version of the CCNG1 enhancer in HCT116 p53+/+ or p53−/− cells. (***P < 0.001 by one-way ANOVA). (F) The canonical Sp1/KLF family motif sequence in the CCNG1 enhancer as a transcription factor logo compared to the wild-type, G3, or G4 variants. This motif is located within the R2 site of the CCNG1 CRE, between 0 and 20 bp from the p53 response element. (G) phyloP vertebrate conservation of the Sp1/KLF family motif within the wild-type CCNG1 enhancer sequence showing high conservation at the G3 and G4 positions. Normalized enhancer activity for the minimal promoter only (MP), WT, G2 or G3 CCNG1 variants in (H) HCT116 p53+/+ or (I) HCT116 p53−/− cells (**P < 0.01, ****P < 0.0001, by one-way ANOVA).