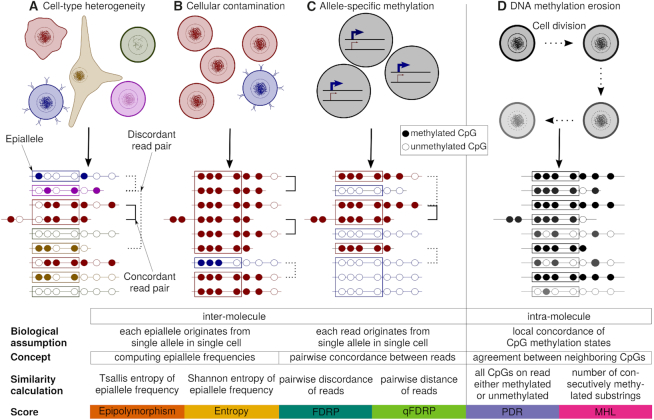
Figure 1.
Sources of WSH and their manifestation in bisulfite sequencing reads. Each line represents a sequencing read. Characteristics of different WSH targeted at quantifying inter-molecule and intra-molecule heterogeneity are outlined below. (A) Cell-type heterogeneity: a sample comprises different cell types with different DNA methylation patterns. (B) Cellular contamination: sample preparation does not yield a pure population of cells of interest. (C) ASM: alleles differ in their DNA methylation states. (D) DNA methylation erosion: cells lose DNA methylation in a stochastic process. Epipolymorphism, Entropy, FDRP and qFDRP describe inter-molecule WSH (scenarios A–C), while PDR and MHL describe intra-molecule WSH (scenario D).