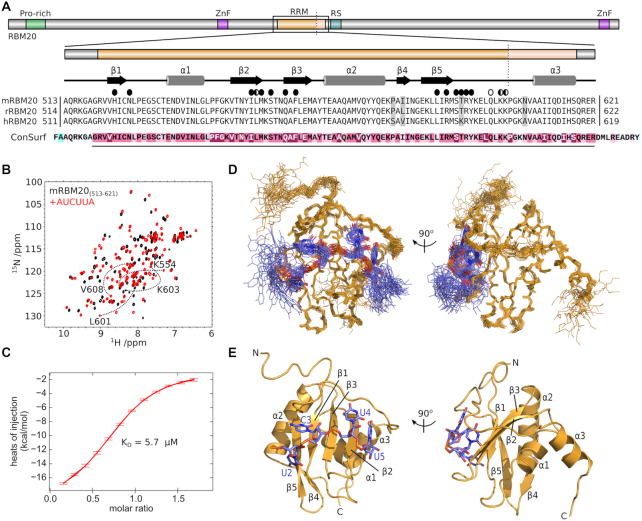
Figure 1.
Recognition of UCUU by the RBM20 RRM domain. (A) Domain composition of RBM20 with expanded details for the RRM domain. The alignment is composed of RBM20 sequences from mouse (mRBM20, UniProt Q3UQS8), rat (rRBM20; UniProt E9PT37) and human (hRBM20; UniProt Q5T481). The vertical dotted line shows the C-terminal end of previous RRM domain constructs. Secondary structure elements from the NMR structure are indicated above the alignment. Residues that contact the RNA by their sidechain and backbone atoms are indicated by full and open circles, respectively. Results from the ConSurf analysis is shown below the alignment, with the region of high conservation indicated with a line, and coloured purple. (B) 15N-HSQC overlay of [15N]mRBM20(513–621) in the absence (black) and presence (red) of 1.2 molar equivalents AUCUUA RNA. Annotated spectra can be found in Supplementary Figure S1. (C) Representative isothermal titration calorimetry (ITC) data for mRBM20(513–621) binding to AUCUUA RNA. (D) Ensemble of 25 lowest energy structures calculated for mRBM20(513–621) (backbone heavy atoms, orange lines) bound to AUCUUA RNA (all heavy atoms, purple lines). (E) Lowest energy structure model for the UCUU motif (all heavy atoms, purple sticks) and mRBM20 (residue 520–617, orange cartoon). RNA bases and protein secondary structure elements are labeled.