Figure 4.
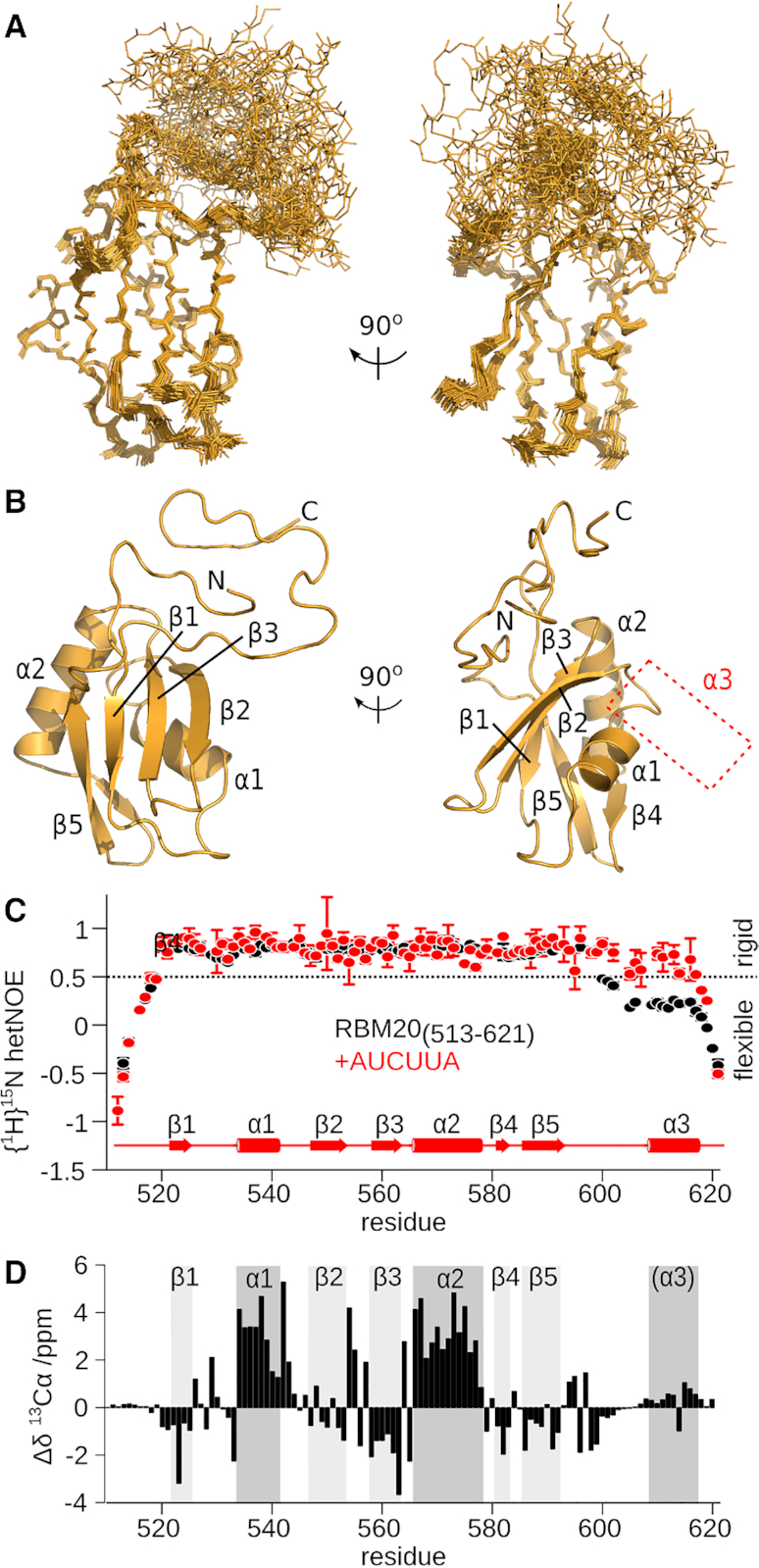
The helix α3 region is disordered in unbound mRBM20(513–621). (A) Ensemble of 25 lowest energy structures calculated for unbound mRBM20(513–621) (backbone heavy atoms, orange lines). (B) Lowest energy structure model for mRBM20 (orange cartoon). Protein secondary structure elements are labelled. The location of α3 in the RNA-bound complex is also indicated. (C) Flexibility of the backbone amides as measured by {1H}15N hetNOE of the unbound (black) and RNA-bound (red) [15N]mRBM20(513–621). The secondary structure elements of RNA-bound mRBM20(513–621) is also shown. Residues with {1H}15N hetNOE values <0.5 are considered to be disordered. (D) 13Cα secondary chemical shift (Δδ) of the unbound mRBM20(513–621) compared to a disordered peptide as predicted by using ncIDP (75). The secondary structure elements of RNA-bound mRBM20(513–621) are indicated by grey boxes.
