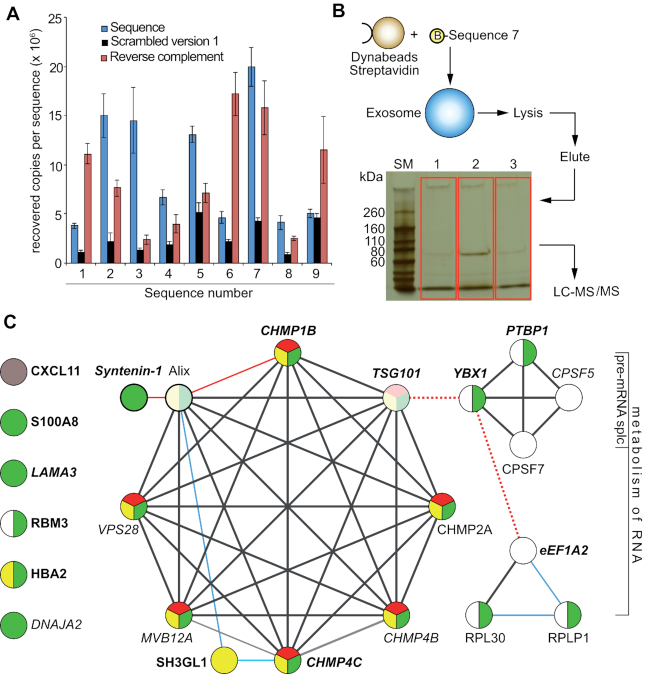
Figure 3.
Target ID. (A) Binding of nine unique sequences (represented as blue dots in 2A) to exosomes from VCaP cells by qPCR; blue bars represent the nine selected sequences. Sequence 7 shows the highest absolute recovery, and was used for target ID; black bars: scrambled versions 1 of the respective sequences; red bars: reverse complement versions of the respective sequences. Error bars: S.D.; n = 3. Binding is expressed as recovered copies per sequence from 2 μg of exosomes as determined by the BCA assay. (B) Affinity purification of target proteins bound to biotinylated Sequence 7 in combination with LC-MS/MS detection identified exosomal binding partners of the aptamer. The gel in red boxes was used for digestion and mass spec analysis. Lane 1: no DNA control; bare beads. Lane 2: pull-down with Sequence 7. Lane 3: pull-down with reverse complement of Sequence 7. (C) Reactome pathway enrichment and STRING 10.5 database analyses of the protein-protein networks of proteins that bound to Sequence 7 with ≥2.5-fold higher abundance than to the Sequence 7RC or to the no oligo bead only control. Black lines: direct or indirect protein-protein associations, edge confidence level high (0.7) to highest (0.9); blue lines: confidence level medium (0.4); red lines: confidence level low (0.15); red dotted lines: interaction not denoted by Reactome or STRING 10.5, but by references (21) for TSG101/YBX1 and (22) for YBX1/eEF1A2 mRNA. Circles represent proteins, protein isoforms, or PTMs produced by a single, protein-coding gene locus. Green circles: proteins associated with exosomes; yellow circles: enriched in the endocytic pathway; red circles: annotated by Reactome or STRING as part of the ESCRT complex. White circles: RNA binding proteins. Pale colored circles: connected through Alix (detected by MS but not necessarily enriched by Sequence 7). Proteins shown in boldface are associated with prostate cancer. CXCL11 (grey circle) is not part of any classification.