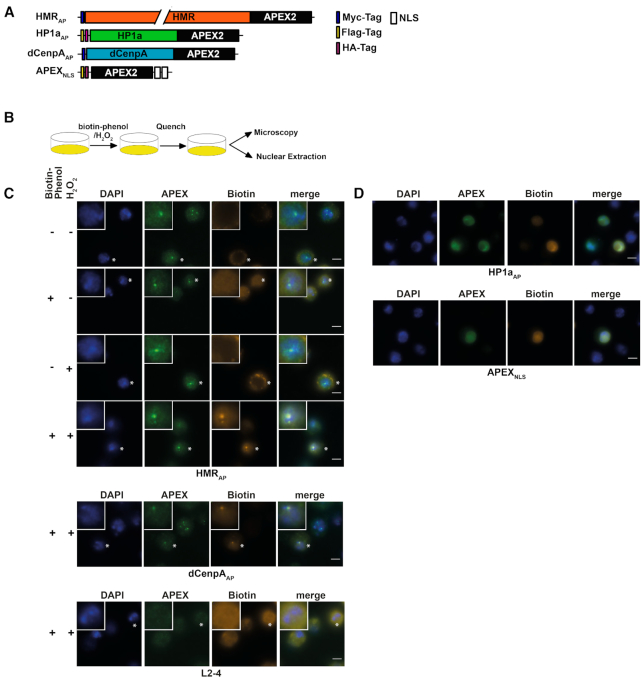
Figure 2.
(A) APEX2 fusion constructs used for proximity based labeling of the chromocenter. (B) Schematic display of a proximity based biotinylation experiment. (C) Biotinylation experiment performed for 1 minute with 5 mM biotin-phenol using the HMRAP cell line (top four panels) in the absence of biotin phenol and hydrogen peroxide, the presence of either biotin phenol or hydrogen peroxide or both reagents. Biotinylation experiment performed for 1 min with 5 mM biotin-phenol using the dCenpAAP cell line (middle panel). The bottom panel shows a treatment of non APEX2 expressing L2–4 cells treated with biotin phenol and hydrogen peroxide. For selected cells (indicated by an asterisk) the inlet shows an approx. 2.3-fold zoom of the nucleus. (D) Proximity based biotinylation of the HP1aAP and the APEX2NLS cell lines using 0.5 mM biotin-phenol. Stainings were performed with anti-APEX2 20H10 antibody and anti-Streptavidin Alexa555. Scale bars represent 5 μm.