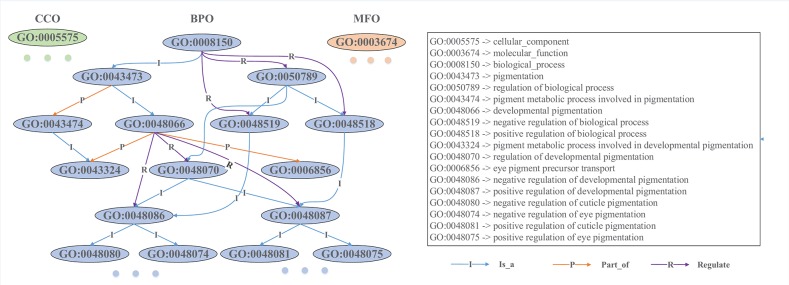
Figure 1.
Snapshot of a directed acyclic graph from Gene Ontology. Each ontological term is represented by an alphanumeric identifier, and its biological function is described by controlled words. These GO terms are hierarchically connected with different types of directed edges. The level of a GO term in the DAG is determined by its furthest distance to the root GO term (“GO:0008150” in BPO, “GO:0005575” in CCO, and “GO:0003674” in MFO). For example, “GO:0048087” is a direct child and also a grandson of “GO:0048066,” and its furthest distance to the root term is 5, while “GO:0006856” is another direct child of “GO:0048066” and its furthest distance to the root is 4, so “GO:0006856” is plotted at a higher level than “GO:0048087”.