Since 1999, dengue outbreaks in the continental United States involving local transmission have occurred only episodically and only in Florida and Texas. In Florida, these episodes appear to be coincident with increased introductions of dengue virus into the region through human travel and migration from countries where the disease is endemic. To date, the U.S. public health response to dengue outbreaks has been largely reactive, and implementation of comprehensive arbovirus surveillance in advance of predictable transmission seasons, which would enable proactive preventative efforts, remains unsupported. The significance of our finding is that it is the first documented report of DENV4 transmission to and maintenance within a local mosquito vector population in the continental United States in the absence of a human case during two consecutive years. Our data suggest that molecular surveillance of mosquito populations in high-risk, high-tourism areas of the United States may enable proactive, targeted vector control before potential arbovirus outbreaks.
KEYWORDS: dengue virus serotype 4, transmission, Aedes aegypti, DENV4, flavivirus, mosquito, arbovirus, surveillance, insect-specific viruses
ABSTRACT
The incidence of locally acquired dengue infections increased during the last decade in the United States, compelling a sustained research effort concerning the dengue mosquito vector, Aedes aegypti, and its microbiome, which has been shown to influence virus transmission success. We examined the “metavirome” of four populations of Aedes aegypti mosquitoes collected in 2016 to 2017 in Manatee County, FL. Unexpectedly, we discovered that dengue virus serotype 4 (DENV4) was circulating in these mosquito populations, representing the first documented case of such a phenomenon in the absence of a local DENV4 human case in this county over a 2-year period. We confirmed that all of the mosquito populations carried the same DENV4 strain, assembled its full genome, validated infection orthogonally by reverse transcriptase PCR, traced the virus origin, estimated the time period of its introduction to the Caribbean region, and explored the viral genetic signatures and mosquito-specific virome associations that potentially mediated DENV4 persistence in mosquitoes. We discuss the significance of prolonged maintenance of the DENV4 infections in A. aegypti that occurred in the absence of a DENV4 human index case in Manatee County with respect to the inability of current surveillance paradigms to detect mosquito vector infections prior to a potential local outbreak.
IMPORTANCE Since 1999, dengue outbreaks in the continental United States involving local transmission have occurred only episodically and only in Florida and Texas. In Florida, these episodes appear to be coincident with increased introductions of dengue virus into the region through human travel and migration from countries where the disease is endemic. To date, the U.S. public health response to dengue outbreaks has been largely reactive, and implementation of comprehensive arbovirus surveillance in advance of predictable transmission seasons, which would enable proactive preventative efforts, remains unsupported. The significance of our finding is that it is the first documented report of DENV4 transmission to and maintenance within a local mosquito vector population in the continental United States in the absence of a human case during two consecutive years. Our data suggest that molecular surveillance of mosquito populations in high-risk, high-tourism areas of the United States may enable proactive, targeted vector control before potential arbovirus outbreaks.
INTRODUCTION
Approximately 40% of the globe is at risk of infection by flaviviruses, such as dengue virus (DENV), an enveloped, single-stranded RNA virus transmitted primarily by Aedes aegypti mosquitoes (1, 2). Since severe disease from DENV infections can manifest as dengue hemorrhagic fever/dengue shock syndrome, DENV establishment in the continental United States is a major concern for public health agencies (1). In the United States, Florida has experienced increases in local DENV transmission since 2009, driven in part by human and pathogen movement (3). A. aegypti is endemic throughout subtropical Florida, and the vector population has resurged recently, following its near-displacement by A. albopictus (4). Autochthonous DENV infection occurs sporadically, primarily in southern Florida, with limited local cases elsewhere in the state (3).
Recently, reports have indicated that certain insect-specific viruses (ISVs) can negatively impact or enhance arbovirus (including DENV) infections in insect cells and mosquitoes (5–7). Although the impacts of many ISVs on arboviral competence have yet to be determined, the evidence to date clearly indicates that the mosquito virome cannot be ignored and likely influences the risk of autochthonous DENV transmission once the virus is introduced into an area. Therefore, we conducted a metaviromic study of A. aegypti adult F1 female (first-generation) mosquitoes raised from eggs collected from ovitraps in 2016 to 2017 from Manatee County to assess the presence of any potentially influential ISVs in local mosquito populations outside southern Florida. Although no indexed human case of DENV4 was reported during 2016 to 2017 in the county, we detected and sequenced DENV4, which was maintained vertically for one generation (since the adults were raised in the laboratory from field-caught A. aegypti eggs), in four mosquito populations from Florida’s Gulf Coast. We followed up this unexpected finding with genetic analyses to determine the DENV4 strain’s likely location of origin, to assess the time frame of virus introduction, and to investigate strain-specific mutations that may have potentially enabled adaptation to and/or persistence within local mosquito populations.
RESULTS
DENV4 and ISVs in A. aegypti mosquitoes from Manatee County, FL.
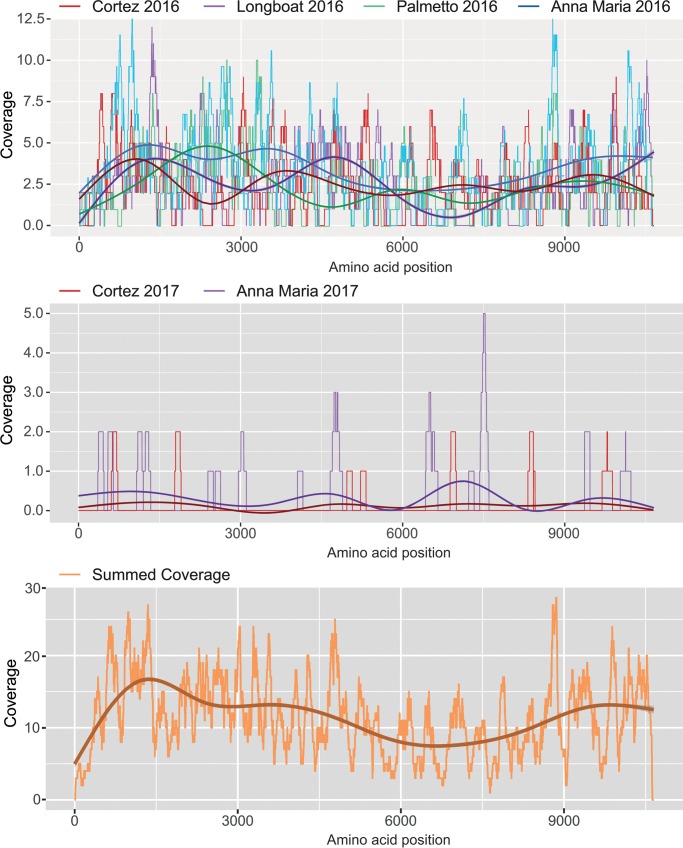
Our metaviromic analysis of female A. aegypti mosquitoes detected DENV4 alongside several ISVs in four sites in 2016 and in only the Anna Maria and Cortez sites in 2017 (Fig. 1). A full DENV4 genome (GenBank accession no. MN192436) was constructed with overall genome coverage of ∼11× across the reads (Fig. 2). We observed that the 2017 DENV4 signal was much lower than the 2016 DENV4 signal for Anna Maria and Cortez (Fig. 3; also, see Fig. S1 in the supplemental material), and although Palmetto had the highest proportion of 2016 reads, this signal was virtually absent in 2017. To confirm DENV4 infection, we amplified and confirmed by direct sequencing the NS2A DENV4 amplicon for 2016 Longboat and Palmetto mosquito samples. Cumulatively, the data indicate that the drop in DENV4 signal relative to the “metavirome” from 2016 to 2017 was statistically significant (effect size of −2.026; P = 0.035).
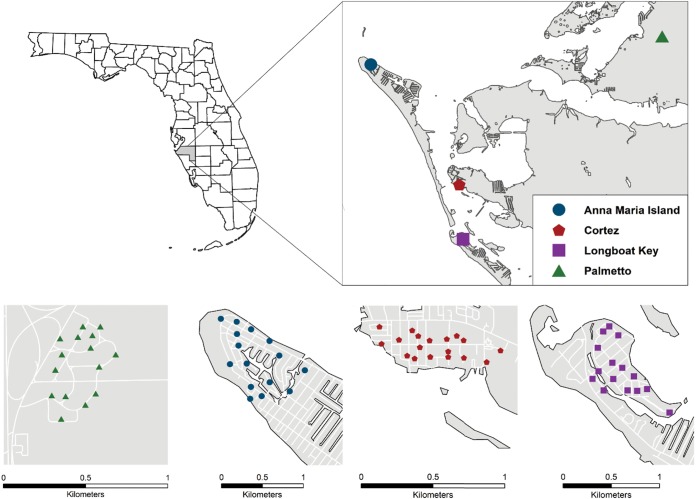
FIG 1.
Locations of ovitraps in four different locations in Manatee County. The collection sites include the city’s subdivisions of Palmetto, Cortez, Ana Maria Island, and Longboat Key.
FIG 2.
Mapping of RNASeq reads on the DENV4 genome. Coverage plots for DENV4 genome readings are shown from top to bottom in the graph panels. Coverage values across the genome for collection site/year combinations are shown as indicated. Coverage is depicted on each y axis and amino acid position on the x axes. The smoothed central lines on the graphs indicate median values.
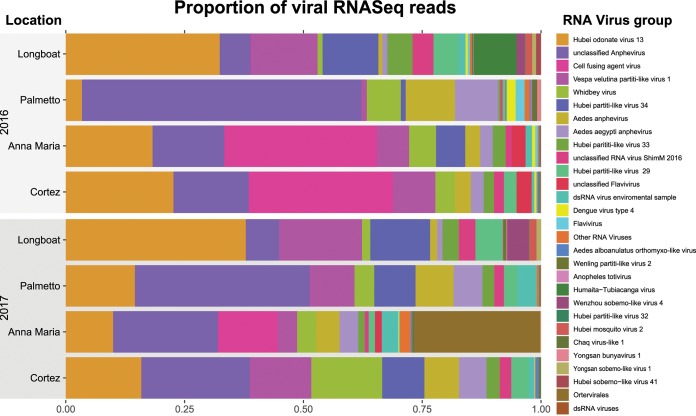
FIG 3.
Metaviromic analysis of Aedes aegypti mosquito populations from Manatee County, FL. Relative abundances of the reads corresponding to RNA viruses in the 8 metagenomes are shown. The proportion of the subcomposition is summarized at the species level for most viruses; however, some viruses were classified at higher levels if species could not be determined by the lowest common ancestor method. dsRNA, double-stranded RNA.
RNASeq read-proportion analyses. To analyze the read abundance of the RNASeq assay conducted on the mosquito samples from Manatee County, the ratio of DENV4-mapped reads to the total number of reads mapped for each site and year pool was calculated. The number of reads that mapped specifically to DENV4 was divided by the total number of reads mapped, the resultant values being shown above the bars on the graph. Proportion values are shown on the y axis of the graph. Download FIG S1, PDF file, 0.6 MB (598.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Manatee County A. aegypti RNA metavirome profile (Fig. 3) indicated abundances of Partitiviridae, Anphevirus, Whidbey virus, and cell fusing agent virus (CFAV). Partitiviridae are known to primarily infect plants, protozoa, and fungi, but all the abundant groups in the metavirome have previously been detected in mosquitoes. We noted that the highest levels of CFAV (Anna Maria and Cortez sites) in 2016 were associated with DENV4 persistence into 2017 (P = 0.07109; R2 = 0.7943). Additionally, Anphevirus signals were notably abundant in the Palmetto samples in 2016 and 2017, coincident with DENV4 signal loss in Palmetto in 2017.
DENV4 phylogenetic and molecular clock analyses.
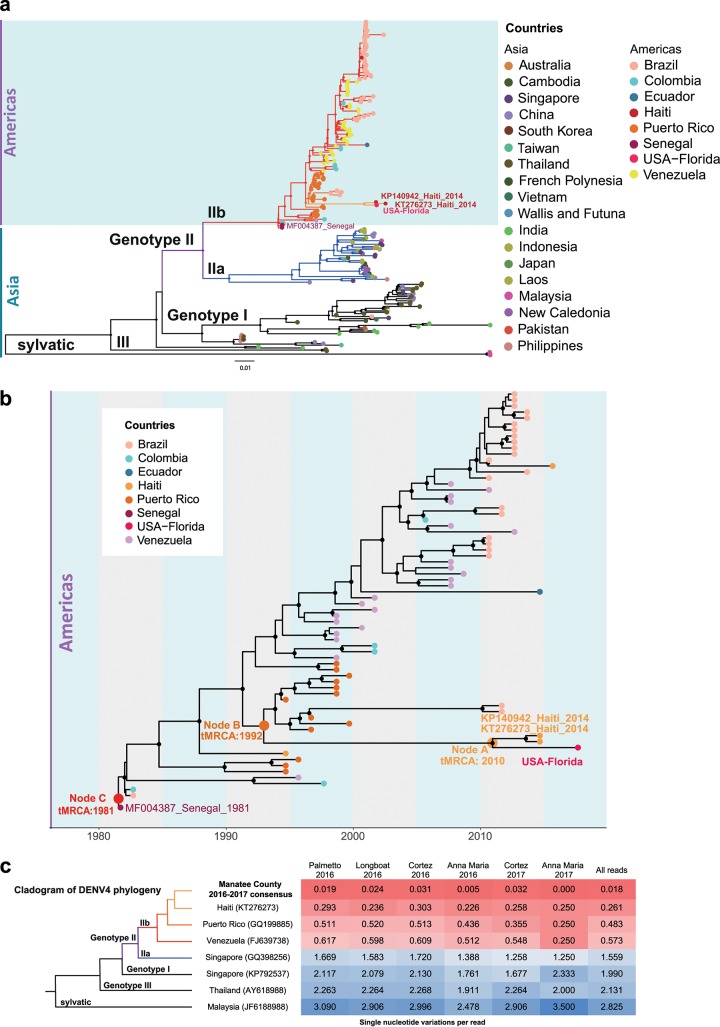
After analyzing the metavirome, we investigated the genome of the DENV4 strain to determine its likely source and to assess the potential time frame of introduction into Florida. Our first analysis confirmed the phylogenetic signal and absence of nucleotide substitution saturation (Fig. S2a and b). We subsequently explored Manatee County DENV4’s phylogeny with a 234-genome DENV4 data set constructed from GenBank sequences (see Table S1 in the supplemental material) by maximum likelihood (ML) phylogenetic inference (Fig. 4a). The ML phylogeny showed three clades: two Asian clades and one American clade with two Senegalese strains (MF00438; GenBank accession no. KF907503) and one Thai strain (GenBank accession no. KM190936) at the base (Fig. 4a). Manatee County DENV4 can be classified as DENV4 genotype IIb. The DENV4 genome obtained in Florida clustered most closely with two Haitian isolates from 2014 (GenBank accession no. KT276273 and KP140942) and a cluster of Puerto Rican isolates (Fig. 4a). Further back in time, a Haitian isolate (GenBank accession no. JF262782) collected 20 years earlier also clustered with the Manatee County-associated clade (Fig. 4a).
FIG 4.
Phylogenetic and phylodynamic analyses of Manatee County DENV4. (a) Maximum likelihood phylogenetic analysis of DENV4 full-genome sequences. The ML tree was obtained using IQ-TREE software. Diamonds at splits along the branches indicate strong statistical support along the branches defined by ultrafast bootstrap values of >90 (34). Tips are labeled and colored based on country of origin. (b) Bayesian phylodynamic reconstruction of DENV4 genotype IIb strains. The time-scaled phylogenetic maximum clade credibility tree was inferred using relaxed clock and constant demographic priors implemented in BEAST v1.8.4. Circles represent branches supported by posterior probability values of >0.90. Tips are colored based on location of origin. Node A (time of the most recent common ancestor [tMRCA] 2010), node B (tMRCA 1992), and node C (tMRCA 1981) are labeled on the branches. (c) SNVs/read per collection site/year combination of mosquitoes with significant detection by viral RNASeq in comparison to various reference genomes shown as a distance matrix. The total numbers of SNVs were normalized by the total numbers of reads from each sample. Cell values refer to the SNV/read ratios of every sample (columns) compared to every representative sequence (rows). Cells are color coded in the matrix as follows: red, 0.0 SNVs/read; white, 1.5 SNVs/read; blue, >3 SNVs/read.
Assessment of phylogenetic quality for DENV4 strains. (a and c) Evaluation of the presence of phylogenetic signal satisfying resolved phylogenetic relationships among sequences was performed by likelihood mapping (IQ-TREE: http://www.iqtree.org/), which estimates the likelihood of each of the three possible tree topologies for each group of four sequences (quartet) in the data set using the best-fit nucleotide substitution model chosen according to the Bayesian information criterion (BIC). Quartets were considered resolved when the three likelihoods were significantly different (phylogenetic signal) and were considered unresolved, or partially resolved, when two likelihood values or all three likelihood values were not significantly different (phylogenetic noise). Percentages shown within each triangle indicate the proportion of resolved quartets (in the three corner areas) and the proportion of partially resolved (side areas) or unresolved (center) quartets. Extensive simulation studies have shown that side/center areas, including <40% of the unresolved quartets, can be considered robust in terms of phylogenetic signal (K. Strimmer, A. von Haeseler, Proc Natl Acad Sci U S A 94:6815–6819, 1997, https://doi.org/10.1073/pnas.94.13.6815; K. Strimmer, A. von Haeseler, Mol Biol Evol 13:964–969, 1996, https://doi.org/10.1093/oxfordjournals.molbev.a025664). (b and d) Substitution saturation, which decreases the phylogenetic information contained in the sequences, was assessed using DAMBE7 (http://dambe.bio.uottawa.ca/DAMBE/dambe.aspx) by plotting pairwise nucleotide (blue) transition (s) and (green) transversion (v) substitutions (y axis) versus pairwise genetic distances (x axis) determined with the Tamura and Nei 1993 (TN93) nucleotide substitution model (K. Tamura, M. Nei, Mol Biol Evol 10:512-526, 1993, https://doi.org/10.1093/oxfordjournals.molbev.a040023). Download FIG S2, PDF file, 0.8 MB (857.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Molecular clock and demographic prior model testing for phylogenetic analyses. Download Table S1, PDF file, 0.6 MB (663.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To estimate the most recent common ancestor (MRCA) for DENV4 entry into Manatee County, FL, as well as the date of divergence of the strain with Haitian isolates, we performed a molecular clock analysis using a Bayesian evolutionary framework on a reduced data set that included only the “Americas clade” (8). We assessed the phylogenetic signal and the absence of nucleotide substitution saturation (Fig. S2c and d) first and then the temporal signal alone (Fig. S3). In the maximum clade credibility (MCC) tree, Manatee County DENV4 clustered with the Haitian isolates from 2014 (node A posterior probability [PP] > 0.9) (Fig. 4b). The MCC phylogeny showed that the time of the MRCA (tMRCA) for the DENV4 Manatee County isolate and Haitian isolates was 2010 (node A in Fig. 4b). This 95% high posterior density (HPD) interval for this tMRCA suggests that DENV4 may have entered Manatee County sometime between 2006 and 2013. For node B (Fig. 4b), the tMRCA of 1992 with a 95% HPD interval of 1901 to 1994 indicated that the Manatee County and 2014 Haitian strains diverged from the 1994 Haitian DENV4 (GenBank accession no. JF262782), almost a decade before its arrival to Florida. However, strain divergence may have occurred in Haiti and was not necessarily precipitated by its introduction into Manatee County. Therefore, the introduction time frame could have been more recent than the estimated tMRCA.
Assessment of temporal signal for DENV4 strains. The plot represents regression analysis of root-to-tip genetic distances assessed using TempEst v1.5. The positive slope (R2 = 0.7135) indicates the presence of temporal signal for the dataset. Download FIG S3, PDF file, 0.5 MB (502.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DENV4 SNVs/read analyses.
Next, we examined the Manatee County DENV4 genome sequences to compare strain variations between years and to identify mutations unique to the strain that potentially enabled local adaptation to and/or persistence in local mosquito populations. Following the MCC phylogenetic analysis, site-specific reads from mosquito populations in Manatee County were analyzed for single-nucleotide variations (SNVs) by determining the number of SNVs/read against the Manatee County consensus genome and other global DENV4 genomes (Fig. 4c). SNV/read values showed only 22 SNVs across the 11,650-nucleotide Manatee County genome against all reads. SNVs were more substantial per read in the other DENV4 genomes. This indicates the likely persistence of a single strain of DENV4 in Manatee County during the 2016–2017 transmission seasons.
Signatures of Manatee County DENV4 adaptation.
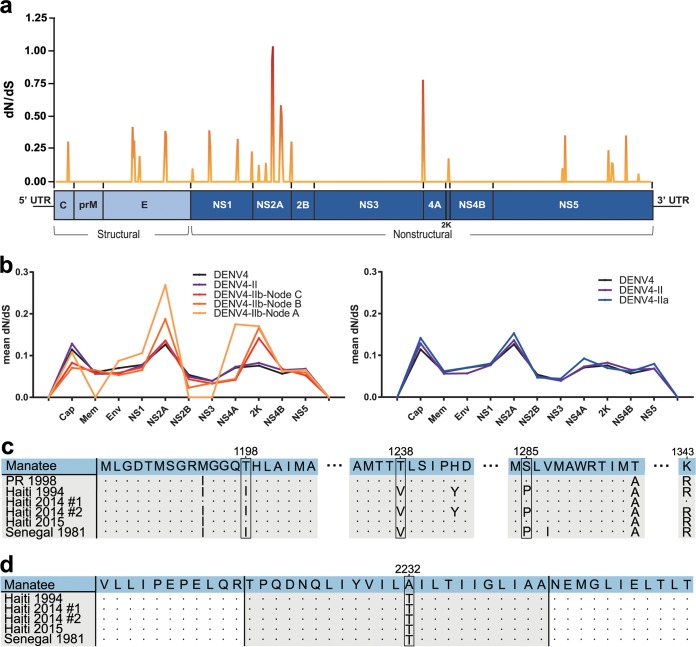
We then explored selective pressures on the Manatee County DENV4 strain’s coding sequence that may be functionally important with respect to transmission and persistence of DENV4 in Floridian aegypti. The DENV4 genome has 5′ and 3′ untranslated regions (UTRs) flanking eight protein-coding genes (encoding nonstructural [NS] protein 1, NS2A, NS2B, NS3, NS4A, the 2K peptide, NS4B, and NS5) (Fig. 5a). The protein-coding regions of Manatee County DENV4 were compared to those of four Haitian DENV4 genomes from 1994 to 2015 and a 1981 Senegalese DENV4 genome. These were analyzed for all amino acid substitutions between strains, and analysis of the ratio of nonsynonymous to synonymous evolutionary changes (dN/dS) was conducted comparing the Senegalese DENV4 genome with the Manatee County DENV4 genome (Fig. 5a; see also Table S2). The highest proportions of amino acid substitutions were seen in NS2A and the 2K peptide; simultaneously, the highest dN/dS values occurred for the NS2A gene, to a point of weak positive selection (dN/dS > 1) that covered a V1238T mutation discussed further here. We then calculated dN/dS data for all DENV4 genomes and for genotype II, genotype IIa, and genotype IIb with all sequences available, as well as within the Haiti-Florida and Haiti-Florida-Puerto-Rico clades (Fig. 5b). Purifying selection, which occurs when nonsynonymous mutations are deleterious, dominated, but we found weaker purifying selection in NS2A and 2K peptide genes, correlating to the Manatee County-to-Senegal dN/dS analysis conducted previously. Values of dN/dS for these genes increased relative to those for flanking genes for genotype IIb and Caribbean/Florida-specific groups as well (Fig. 5b).
FIG 5.
DENV4 amino acid analyses. (a) dN/dS analysis conducted for the whole coding region of the DENV4 Manatee County genome versus the Senegalese genome from 1981 (MF004387.1). The analysis was conducted utilizing full-genome coding sequences in JCoDA with a sliding window analysis and a window size of 10. A small genome schematic is placed below the graph. Red indicates lower dN/dS values; orange indicates higher values; color shades between red and orange indicate intermediate values. (b) (Left) Using all available sequences, a dN/dS comparison was conducted to calculate mean ratio values within DENV4 for all genotypes, DENV4 genotype II, DENV4 genotype IIb, DENV4 Florida-American-Caribbean-specific clades, and DENV4-Florida-Caribbean-specific clades (corresponding to nodes C, B, and A as indicated in Fig. 4b). (Right) A comparison between all available DENV4 genome sequences, DENV4 genotype II, and DENV4 genotype IIa is depicted. (c) Amino acid sequence alignment of Manatee County (MN192436), Puerto Rican (AH011951.2), Haitian 1994 (JF262782.1), Haitian 2014 no. 1 (KP140942.1), Haitian 2014 no. 2 (KT276273.1), Haitian 2015 (MK514144.1), and 1981 Senegalese (MF004387.1) genome sequences for the NS2A region previously sequenced in Puerto Rican isolate genomes by Bennet et al. (9). Amino acid positions are numbered at the top of the panel. Key amino acid changes defining the 1998 DENV4 Puerto Rican outbreak in the NS2A gene are highlighted with boxes. The PR 1998 sequence shows some differences from and similarities with the compared sequences for the outbreak’s key mutations. (d) A comparison of 2K peptide (colored in gray) sequences between the Manatee County (MN192436), 1994 Haitian (JF262782.1), Haitian 2014 no. 1 (KP140942.1), Haitian 2014 no. 2 (KT276273.1), Haitian 2015 (MK514144.1), and 1981 Senegalese (MF004387.1) genomes. Uncolored portions of the sequences correlate to portions of NS4A and NS4B.
Amino acid differences between Manatee County, Haitian, and Senegal DENV4 genomes. Download Table S2, PDF file, 0.6 MB (631.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, we further analyzed coding sequences in specific regions of the genome to investigate specific mutations that may have mediated Manatee County DENV4 Floridian entry and persistence. The NS2A gene was analyzed in an alignment comparison between Manatee County (GenBank accession no. MN192436), 1994 Haitian (GenBank accession no. JF262782.1), 2014 Haitian no. 1 (GenBank accession no. KP140942.1), 2014 Haitian no. 2 (GenBank accession no. KT276273.1), 2015 Haitian (GenBank accession no. MK514144.1), and 1981 Senegalese (GenBank accession no. MF004387.1) full genomes and a partial DENV4 genome (GenBank accession no. AH011951.2, Puerto Rico, 1998), with the analysis targeting three mutations that defined the 1998 DENV4 Puerto Rican outbreak (Fig. 5c) (9). The Manatee County DENV4 sequence shares these key mutations with 1998 Puerto Rico, 2014 Haiti no. 1, and 2015 Haiti genomes. Conversely, the 1981 Senegal sequence and the oldest Haitian sequence from 1994 lack these mutations. In a selective pressure analysis utilizing the aforementioned 234-genome assembly, we observed strong background purifying selection with 143 sites that were found to be under episodic negative/purifying selection within the NS2A gene. Episodic diversifying/positive selection (evolutionarily preferred nonsynonymous mutation) was detected in two sites corresponding to amino acids 1238 and 1333; both residues localized to transmembrane segments of the protein. This makes V1238T a mutation of note corresponding to the previous NS2A-associated analysis, detected in different analyses as a point of possible positive selection. The 2K peptide was next analyzed against the four Haitian genomes and the Senegalese genome from the first NS2A-specific analysis (Fig. 5d), and we observed that it had the second highest general rate of nonsynonymous mutations and had a peak of weaker purifying selection (Fig. 5a). There was only one nonsynonymous mutation among the six genomes, which is significant considering the size of the 2K peptide. This was a T2232A mutation present solely in the Manatee County DENV4 sequence.
DENV4 3′ UTR sequence and secondary structure analysis.
To complete our genomic analysis of Manatee County DENV4, we examined the 3′ UTR, as this region and its derivative subgenomic (sRNA) RNA have been implicated in epidemiologic and transmission fitness. Although the DENV4 3′ UTR lacks one of the two flaviviral nuclease-resistant RNAs (fNRs) in domain I compared to other DENV 3′ UTRs (3′ UTRs of DENV serotypes 1 to 3), DENV4 has the same conserved secondary structures in its domain II and domain III: two dumbbells (dumbbell 1 [DB1] and DB2) and a 3′-end stem-loop (3′ SL) (Fig. S4). The 3′ UTR, through structural conformations, can affect viral replication in hosts (10). We noted several transition substitutions in the DENV4 IIb lineage prior its arrival in Florida (node B in Fig. 4b and Fig. S4b). Most of these mapped to either the highly variable region (HVR) or the adenine-rich segments that space functional RNA elements in DENV 3′ UTRs (11). The U10318C substitution in fNR2 (fNR1 present only in other DENV serotypes) and the G10588A substitution on the 3′ SL mapped to base-pairing positions. Manatee County DENV4 also underwent a rare transversion (A10478U) in a conserved position in DB2. This substitution favors formation of a new base pair in the structure of DB2. Additionally, an insertion (10467A) occurred in the adenine-rich segment upstream of DB2; this insertion is common for all lineages.
DENV4 3′ UTR analyses. (a) Alignment with the 3′ UTR of Manatee County DENV4. The 3′ UTR RNA sequence alignment is of DENV4 from Manatee County, DENV4 Haiti 2014 no. 2 (GenBank accession no. KT276273.1), and DENV4 Philippines H241 (GenBank accession no. KR011349.2) genomes. The DENV4 RNA sequence alignment was generated with CLC Sequence Viewer 8.0 (http://resources.qiagenbioinformatics.com/manuals/clcsequenceviewer/current/index.php?manual=Introduction_CLC_Sequence_Viewer.html). Some conserved DENV4 3′ UTRs are designated in black boxes in the figure, including repeated conserved sequence 2 (RCS2), conserved sequence 1 (CS1), conserved sequence 2 (CS2), and a 3′ upstream AUG region (3′ UAR). Different nucleotides are designated with different colors. (b) A diagram of the secondary structure of the DENV4 Manatee County 3′ UTR is depicted with key nucleotides and mutations highlighted and drawn in orange, correlating to node A or node B from Fig. 4B. Key secondary structure regions of the 3′ UTR are shown in black text as follows: dumbbell 1 (DB1) and DB2, pseudoknots 1 to 5 (PK1 to PK5), flavivirus nuclease-resistant RNA (fNR2), and the 3′ stem loop (3′ SL). Download FIG S4, PDF file, 0.8 MB (785KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Our metavirome analysis of A. aegypti from Manatee County has revealed potential insight into, and new possible examples of, human arboviruses and ISV relationships in a state prone to autochthonous flavivirus transmission, considered in light of previous research on such interviral relationships. The observed drop in DENV4 relative to the mosquito virome (ISVs) between 2016 and 2017 was statistically significant (P = 0.035), opening the possibility that the ISV profile for individual mosquitoes may influence persistence of DENV4 in site-specific mosquito populations within the surveyed area. Note that our analysis was conducted on pools of mosquitoes, which limits the conclusions that can be drawn in this study versus an analysis conducted on individual mosquitoes and interviral dynamics. For example, although Anphevirus (described in our analyses) has been shown to reduce DENV titers in vitro during coinfections, further analysis performed using individual mosquitoes of this specific dynamic is necessary to fully determine whether Anphevirus coinfection (occurring singly or concomitantly with other ISVs) can mediate multigenerational dengue virus persistence in mosquitoes (12).
With respect to the role of natural infections by insect-specific flaviviruses in the proliferation of pathogenic arboviruses carried by different mosquito vector species, current knowledge is uncertain. A mosquito-specific flavivirus that we detected that is known as cell fusing agent virus (CFAV) is of particular interest. Coinfection studies performed in vitro with DENV2 and CFAV resulted in enhanced proliferation in both (13). The observed correlation between persistence of DENV4 infection into 2017 in Anna Maria and Cortez mosquitoes and CFAV abundance in 2016 (Fig. 3) may provide an example of the dynamics described previously by Zhang et al., showing the enhanced replication of the two viruses (13). An important caveat is that the research reported by Zhang et al. was conducted in vitro. Conversely, Baidaliuk et al. demonstrated in vivo amplification-restrictive interactions between CFAV and DENV1 (7). How interactions between DENV4 genotype, mosquito genotype, and CFAV genotype ultimately influence the vector competence of Floridian A. aegypti mosquitoes remains to be determined. The observed metavirome patterns set the stage for follow-up studies to characterize the precise nature of ISV-DENV-mosquito interactions viz. vector competence.
The absence of an index human DENV4 case does not preclude the possibility that DENV4 was transmitted locally. Up to 88% of primary DENV infections are asymptomatic, with DENV4 being widely understood to cause primarily subclinical infections (14, 15). Importantly, clinically inapparent infections could contribute to 84% of DENV transmission events through mosquitoes, so the threat of local transmission cannot be ruled out (14). However, it is noteworthy that DENV4 was detected in adult female mosquitoes reared from wild-captured eggs, implicating transovarial transmission (TOT) in local A. aegypti as has been shown for DENV1 in Key West, FL (16). However, since the DENV4 signal measured in 2017 was lower than that measured in 2016, with two sites losing DENV4 prevalence, if it had played a role in maintaining DENV4 in Manatee County mosquitoes, TOT alone might have been insufficient to maintain DENV4 from 2016 to 2017. At present, vertical transmission remains only a possibility, especially since this phenomenon has not been fully described outside laboratories. Furthermore, we suspect that despite Manatee County DENV4’s divergence from Haitian strains sometime between 2006 and 2013, it likely did not enter Manatee County until 2014 or after, given its similarity to the 2014–2015 Haitian DENV4 isolates and the fact that TOT is an inefficient process. A recent review evaluating the influence of TOT in DENV epidemiology concluded that the current body of research suggests that vertical transmission is likely to be insufficient to represent an independent mechanism of DENV maintenance (17). Tertiary mechanisms, beyond ISV composition profile and TOT, could include inapparent human-mosquito infection cycles during the summer transmission (mosquito) season, which may have also contributed to DENV4 persistence in Manatee County aegypti. The exact mechanisms of maintenance in mosquitoes and proof of local transmission are difficult to elucidate at this juncture, considering that all mosquito samples were processed for rRNA-depleted total RNA sequencing (RNASeq) and reverse-transcription PCR (RT-PCR) (i.e., no live virus can be isolated). Importantly, a comprehensive serological survey with subsequent confirmation by gold-standard neutralization assay of the population from the four sample collection sites was not possible within the estimated mean half-life of detectable anti-DENV4 virion IgM or IgG. This limitation was unavoidable since (i) the complete viral genome assembly and orthogonal confirmation occurred more than 2 years following the initial mosquito collections and (ii) there are significant confounders and logistical obstacles (well outside the current scope of the study) that complicate working with transient worker and migrant communities in the sampled area. However, the data representing complete assembly and persistence over 2 years of an individual strain of DENV4, which is supported by results from orthogonal analytical approaches, remain provocative and reveal an unappreciated ecological process for DENV4 transmission in a nonendemic setting.
Tracking and predicting movement and introduction of arbovirus into the United States, especially into Florida, can potentially lead to proactive efforts for increased monitoring and vector control at critical points of introduction into the state. DENV4 has been reported throughout the Caribbean, especially in Puerto Rico and Haiti and, more recently, in Cuba (18). Florida has the largest populations of people of Puerto Rican, Haitian, and Cuban origin and descent in the United States, and there are ongoing efforts to develop effective “sentinel” surveillance programs that can prepare Florida to deal with potential local arbovirus transmission. As expected, our analysis suggests a Caribbean origin for the Manatee County isolate due to movements of DENV4 into Florida from Haiti and, preceding that, into Haiti from Puerto Rico. These results agree with previous findings depicting the Caribbean as a hot spot for arboviral spread in the Americas (18–20). Diversifying selective pressure in the NS2A gene and the 2K peptide (Fig. 5a and b) experienced by American/Caribbean DENV4 may have contributed to the fixation of mutations driving the adaptation of DENV4 to local infections of human and mosquito populations. NS2A mutations that characterized the 1998 DENV4 outbreak in Puerto Rico are conserved between the Manatee County, Puerto Rican, and two Haitian genomes (GenBank accession no. JF262782.1 and KT276273.1) (Fig. 5c) (9). The 1981 Senegalese strain, the strain clustering closest to the Manatee County strain isolated outside the Americas (Fig. 4a and b), shares none of these mutations with Manatee County DENV4. An in-depth understanding of how putative “hallmark” mutations in arboviruses can lead to increased local aegypti mosquito infections is lacking, compelling further study.
We observed the expected 15-nucleotide deletion (Δ15) in the Manatee County DENV4 3′ UTR (see Fig. S4 in the supplemental material) that is present across all circulating DENV4 strains but absent from the extinct genotype I DENV4 lineage (GQ868594_Philippines_1956). Since the Δ15 deletion maps to the HVR, it does not alter the secondary structures required for subgenomic flaviviral RNA (sfRNA) production. However, the HVR is an adenylate-rich unfolded spacer with poor sequence conservation—where no reliable secondary structure can be predicted, as our previous analyses suggested (11). It has been speculated that these spacers favor the correct folding of adjacent functional structured RNA elements. The deletion might change the rate of folding of the downstream functional structured RNA and thus might alter sfRNA production levels. Clearly, a closer molecular exploration of the exact role of this Δ15 deletion is needed.
The potential implications of our findings are intriguing, especially considering that arboviral surveillance of mosquito populations during the extended Florida mosquito season (April to October) is limited. To our knowledge, this is the first reported characterization of a DENV4 infection in native mosquito populations in Florida in the absence of an index human case across 2 years in a specific county. These data highlight the importance of knowing when and where arboviruses are introduced and point to the potential benefit of surveilling local mosquito populations for arbovirus infections prior to an outbreak. Given the increasing number of travel-related arbovirus introductions into Florida alone and the risk of local establishment in the state, we expect that while our report is seminal, it likely represents the tip of the iceberg. Furthermore, in 2019, 16 cases of locally acquired DENV were reported for the state, including an area along the West Central Florida Gulf Coast (21). Among the 335 travel-associated cases in 2019, DENV1 (n = 63), DENV2 (n = 235), and DENV3 (n = 31) serotypes were identified by PCR from 329 samples. DENV serotypes for the local cases are reasonably predicted to mirror the geographical distribution of serotypes for the travel-associated cases. If these data and our own findings are any indication, the number of “under-the-radar” arbovirus infections of mosquito populations in migration hot spots across the state (and perhaps across other states and regions around the globe) remains significantly underestimated.
MATERIALS AND METHODS
Mosquito sample preparation and viral RNASeq.
To avoid cross contamination of genetic material by sympatric A. albopictus in the area and to ensure the preparation of sufficient quantities of RNA extracted from only pristine samples of A. aegypti nulliparous females, we elected not to use adult traps. As such, eggs were collected in ovitraps in 2016 and 2017 (15 May 2016 and 19 June 2017) from four Manatee County sites (Fig. 1). To avoid cross contamination of mosquito viromes, each year eggs from each site were hatched independently in distilled water, reared to adulthood, identified by species, and then frozen. Twenty female mosquitoes were selected from each site, and abdomens were processed as a single pool per site (n = 20/pool) for the four collection sites for a total of eight individual pools. Total RNA was extracted using an AllPrep DNA/RNA minikit (Qiagen), and rRNA was depleted using a NEBNext rRNA depletion kit (New England BioLabs). A NEBNext Ultra II directional RNA library preparation kit (New England BioLabs) was used to prepare shotgun metagenomics libraries. Reverse-transcribed RNA libraries were sequenced using a HiSeq 3000 instrument (Illumina) in 2 × 101 run mode. The data were deposited into the NCBI Sequence Read Archive and Biosample archive under BioProject PRJNA547758.
Initial assembly and metavirome analysis.
BBduk (version 37.75; https://sourceforge.net/projects/bbmap/) was used to trim adaptor sequences and remove contaminants. A. aegypti sequences were removed using BBsplit (https://sourceforge.net/projects/bbmap/) and the A. aegypti Liverpool genome (AaegL5.1). Nonmosquito reads were assembled using Spades (3.11.1) in metagenomics mode (22). For each contig, a local similarity search in protein space was run using Diamond (0.9.17) against the NCBI NR (National Center for Biotechnology Information nonredundant) sequence database (23). Reads were mapped against assemblies using Bowtie (2.3.4.1) and were then sorted/indexed using Samtools (1.4.1) (24, 25). Megan 6 was used to assign contigs and read counts to the lowest common ancestor (LCA; the lowest common ancestor on a phylogenetic tree if situated vertically, making the LCA the nearest common ancestor) and to view viral contigs (26). To estimate microbial community abundance, Diamond (0.9.17) was used to search reads against the NCBI NR database, Megan 6 was used to assign read counts to the LCA, and R (3.6.0) package Compositions (1.40-2) was used to create a subcomposition of RNA that was used to quantify RNA virus taxon abundance by site/year (e.g., Palmetto 2016 and Palmetto 2017) from the body of overall compositional RNA data in each site/year data set (Fig. 3) (23, 26, 27). Compositional count data from the Megan LCA classification were assessed by the use of ALDEx2 to estimate the statistical significance of the change in DENV4 reads from 2016 to 2017 (26, 28, 29). ALDEx2 uses a Dirichlet multinomial Monte Carlo simulation to estimate the variance of the centered log ratio (CLR) values for taxa among the reads (28, 29). Using the variance of the CLR, ALDEx2 computes P values using Welch’s t test and returns an effect size (CLR/variance) for the estimate (28, 29). For a determination of the statistical significance of the observed decrease in cell fusing agent virus (CFAV) reads from 2016 to 2017, a linear regression fitted to the CLRs of the Anna Maria and Cortez site DENV4 reads in 2016 and 2017 was utilized to yield an R2 value and a P value to describe the trend.
DENV4 refinement and genome-closing assembly.
Two contigs covering most of the genome with a small gap were obtained. To create a closed genome, a data set of genomes for DENV1, DENV2, DENV3, and DENV4 (GenBank accession no. NC_001477.1, NC_001474.2, NC_001475.2, and NC_002640.1) and the two assembled contigs were used. We selected reads sharing a 31-mer with the data set using BBduk (https://sourceforge.net/projects/bbmap/), followed by assembly with Spades in meta mode and classification using Diamond for a complete DENV4 genome (22, 23). Read mapping performed with Bowtie revealed incorrect bases near the 3′ end, which were manually corrected (24). The genome was annotated using the Genome Annotation Transfer Utility from the Virus Pathogen Database and Analysis Resource (ViPR) (30, 31).
Phylogenetic and molecular clock analyses.
Two hundred thirty-four DENV4 genome sequences from GenBank (see Table S1 in the supplemental material) were aligned using MAFFT version 7.407 with the L-INS-I method (32, 33). IQ-TREE software was used to evaluate phylogenetic signal in the genomes by likelihood mapping and to infer maximum likelihood (ML) phylogeny based on the best-fit model according to the Bayesian Information Criterion (BIC) (34–36). Statistical robustness for internal branching order was assessed by Ultrafast Bootstrap (BB) Approximation (2,000 replicates), and strong statistical support was defined as represented by BB values of >90% (37).
To estimate when DENV4 entered Florida, we used 145 strains, including all isolates from the Americas, related Asian and African isolates, and randomly reduced oversampled Brazilian isolates. The strains in this data set were not recombinant, as assessed by scanning the alignments for possible recombination points using the RDP, GENECONV, MaxChi, CHIMAERA, and 3Seq algorithms implemented in RDP4 software (available from http://web.cbio.uct.ac.za/~darren/rdp.html) (38). Determinations of correlations between root-to-tip genetic divergence and date of sampling were conducted to assess clock signal before Bayesian phylodynamic analysis (39). Time-scaled trees were reconstructed using the Bayesian phylodynamic inference framework in BEAST v.1.8.4 (40, 41). Markov chain Monte Carlo (MCMC) samplers were run for 200/250 million generations to ensure Markov chain mixing, assessed by calculating the effective sampling size (ESS) of parameter estimates. The HKY substitution model was used with empirical base frequencies and gamma distributions of site-specific rate heterogeneity (42). The fit of strict versus relaxed uncorrelated molecular clock models and constant size versus Bayesian Skyline Plot demographic models were tested (8). Marginal likelihood estimates (MLE) for Bayesian model testing were obtained using path sampling (PS) and stepping-stone sampling (SS) methods (43, 44). The best model consisted of a strict clock and a constant demographic size. The maximum clade credibility tree was inferred from the posterior distribution of trees using TreeAnnotator, specifying a burn-in of 10% and median node heights, and was then edited graphically in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/), alongside ggtree, available in R (45).
Single-nucleotide variation analyses.
The viral RNA sequencing reads were mapped onto the complete genome of seven DENV4 strains. These strains represent all the known DENV4 lineages (accession numbers are provided in Fig. 4c). We also mapped the reads onto the assembled Manatee County DENV4 full genome. The read mapping was performed using the Geneious platform (Geneious Prime version 2019.2.1) and the “map to reference” function with standard settings (Mapper: Geneious; Sensitivity: Highest Sensitivity/Slow; Fine tuning: Iterate up to 5 times; no trim before mapping). The single-nucleotide variation quantification was performed in the same platform using the “find Variation/SNV” function under default settings.
DENV4 genetic analyses.
From the alignment of the 234 DENV4 genomes, sequences corresponding to the NS2A gene were extracted to investigate selection pressure and mutations that potentially influenced adaptation to and/or persistence in mosquito populations. Comparative selection and mutation analyses revealed NS2A to be a relatively strong region of potential selection for the Manatee County genome. HyPhy algorithms were used to estimate nonsynonymous (dN) to synonymous (dS) codon substitution rate ratios (ω), with ω values of <1 indicating purifying/negative selection and ω values of >1 indicating diversifying/positive selection (46, 47). Fast, unconstrained Bayesian approximation (FUBAR) was used for inferring pervasive selection and the mixed-effects model of evolution (MEME) to identify episodic selection (48, 49). Sites were considered to have experienced diversifying/positive or purifying/negative selective pressure based on posterior probability (PP) values of >0.90 for FUBAR and likelihood ratio test result of ≤0.05 for MEME.
To elucidate influential mutations in the Manatee County DENV4 genome that potentially enabled persistence in the local mosquito population, a dN/dS analysis of the Manatee County DENV4 against the closely related but geographically distant 1981 Senegalese DENV4 (GenBank accession no. MF004387.1) was conducted using JCoDA with default settings, a 10-bp sliding window, and a jump value of 5 (50). To further assess the selective pressure throughout coding sequences in the DENV4 lineage that established transmission in Manatee County, we implemented a single-likelihood ancestor counting (SLAC) method using the DataMonkey 2.0 Web application (51, 52). That application combines maximum likelihood (ML) and counting approaches to infer nonsynonymous (dN) and synonymous (dS) substitution rates on a site-by-site basis for the different DENV4 coding alignments and corresponding DENV4 phylogeny. The measurements were performed on different alignments that included all strains, only genotype II strains, only clade IIa or IIb strains, or only strains that are closely related to the DENV4 Manatee County strain (multiple alignments of DENV4 coding sequences are available as a Mendeley data set). NS2A and 2K peptide genes were individually aligned and inspected in closely related DENV4 strains (1994 Haitian [JF262782.1], 2014 Haitian no. 1 [KP140942.1], 2014 Haitian no. 2 [KT276273.1], 2015 Haitian [MK514144.1], and 1981 Senegalese [MF004387.1] genomes) and the Manatee County DENV4 strain for mutations to identify possible signals of adaptation of Manatee County DENV4 to Floridian A. aegypti.
Data availability.
Viral RNASeq read data are available in the NCBI Sequence Read Archive and Biosample archive under BioProject PRJNA547758. Genome sequence data for the Manatee County sequence are available in NCBI’s GenBank database (GenBank accession no. MN192436), and reference sequences are available in the GenBank database under the accession numbers provided in the text. Multiple coding DENV4 sequence alignments from the dN/dS analyses and alignments for the RNA secondary structure model in Fig. S4 in the supplemental material are available with relevant accession numbers in a Mendeley data set (https://data.mendeley.com/datasets/kwszjp63rb/draft?a=e11f9b80-bcfb-443b-918d-3016032ef3bd). The GenBank accession numbers for the sequences compared in the alignments shown in Fig. 5c and d are (from top to bottom in panel c and d, excluding AH011951.2 in panel d for which sequencing data of the strain is not available in the compared region of the genome) MN192436, AH011951.2, JF262782.1, KP140942.1, KT276273.1, MK514144.1, and MF004387.1.
ACKNOWLEDGMENTS
We gratefully acknowledge the support of Carina Blackmore, Danielle Stanek, and Andrea Morrison from the Florida Department of Health, as well as Lisa Conti, Kelly Friend, Davis Daiker, and Adriane Rogers from the Florida Department of Agriculture and Consumer Services for their institutional collaboration with the CDC Southeastern Center of Excellence in Vector Borne Diseases: The Gateway Program. We also thank Heather Coatsworth and Kaci McCoy for useful comments. This research was supported in part by the United States Centers for Disease Control (CDC) Grant 1U01CK000510-03: Southeastern Regional Center of Excellence in Vector-Borne Diseases: The Gateway Program. The CDC had no role in the design of the study, the collection, analysis, and interpretation of data, or in writing the manuscript. Support was also provided by the University of Florida Emerging Pathogens Institute, the University of Florida Preeminence Initiative, and United States Department of Agriculture, Agricultural Research Service, project 6066-21310-005-00-D.
We declare that we have no competing interests.
REFERENCES
- 1.World Health Organization. 2007. Report of the scientific working group meeting on dengue: Geneva, 1–5 October 2006. World Health Organization, Geneva, Switzerland: https://apps.who.int/iris/handle/10665/69787. [Google Scholar]
- 2.Kraemer MU, Sinka ME, Duda KA, Mylne A, Shearer FM, Brady OJ, Messina JP, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Wint GR, Elyazar IR, Teng HJ, Hay SI. 2015. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data 2:150035. doi: 10.1038/sdata.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rey JR. 2014. Dengue in Florida (USA). Insects 5:991–1000. doi: 10.3390/insects5040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiskind MH, Lounibos LP. 2013. Spatial and temporal patterns of abundance of Aedes aegypti L. (Stegomyia aegypti) and Aedes albopictus (Skuse) [Stegomyia albopictus (Skuse)] in southern Florida. Med Vet Entomol 27:421–429. doi: 10.1111/mve.12000. [DOI] [PubMed] [Google Scholar]
- 5.Romo H, Kenney JL, Blitvich BJ, Brault AC. 14 November 2018, posting date Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg Microbes Infect doi: 10.1038/s41426-018-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz MJ, Frydman HM, Connor JH. 2018. Dual insect specific virus infection limits arbovirus replication in Aedes mosquito cells. Virology 518:406–413. doi: 10.1016/j.virol.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baidaliuk A, Miot EF, Lequime S, Moltini-Conclois I, Delaigue F, Dabo S, Dickson LB, Aubry F, Merkling SH, Cao-Lormeau VM, Lambrechts L. 2019. Cell-fusing agent virus reduces arbovirus dissemination in Aedes aegypti mosquitoes. J Virol 93:e00705-19. doi: 10.1128/JVI.00705-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall MD, Woolhouse ME, Rambaut A. 2016. The effects of sampling strategy on the quality of reconstruction of viral population dynamics using Bayesian skyline family coalescent methods: a simulation study. Virus Evol 2:vew003. doi: 10.1093/ve/vew003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. 2003. Selection-driven evolution of emergent dengue virus. Mol Biol Evol 20:1650–1658. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- 10.Gebhard LG, Filomatori CV, Gamarnik AV. 2011. Functional RNA elements in the dengue virus genome. Viruses 3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finol E, Ooi EE. 2019. Evolution of subgenomic RNA shapes dengue virus adaptation and epidemiological fitness. iScience 16:94–105. doi: 10.1016/j.isci.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parry R, Asgari S. 16 August 2018, posting date Aedes anphevirus: an insect-specific virus distributed worldwide in Aedes aegypti mosquitoes that has complex interplays with Wolbachia and dengue virus infection in cells. J Virol doi: 10.1128/JVI.00224-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Asad S, Khromykh AA, Asgari S. 2017. Cell fusing agent virus and dengue virus mutually interact in Aedes aegypti cell lines. Sci Rep 7:6935. doi: 10.1038/s41598-017-07279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ten Bosch QA, Clapham HE, Lambrechts L, Duong V, Buchy P, Althouse BM, Lloyd AL, Waller LA, Morrison AC, Kitron U, Vazquez-Prokopec GM, Scott TW, Perkins TA. 2018. Contributions from the silent majority dominate dengue virus transmission. PLoS Pathog 14:e1006965. doi: 10.1371/journal.ppat.1006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis 181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 16.Buckner EA, Alto BW, Lounibos LP. 2013. Vertical transmission of Key West dengue-1 virus by Aedes aegypti and Aedes albopictus (Diptera: Culicidae) mosquitoes from Florida. J Med Entomol 50:1291–1297. doi: 10.1603/me13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunnill M, Boots M. 2016. How important is vertical transmission of dengue viruses by mosquitoes (Diptera: Culicidae)? J Med Entomol 53:1–19. doi: 10.1093/jme/tjv168. [DOI] [PubMed] [Google Scholar]
- 18.Mavian C, Dulcey M, Munoz O, Salemi M, Vittor AY, Capua I. 2018. Islands as hotspots for emerging mosquito-borne viruses: a one-health perspective. Viruses 11:11. doi: 10.3390/v11010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blohm G, Elbadry MA, Mavian C, Stephenson C, Loeb J, White S, Telisma T, Chavannes S, De Rochar VMB, Salemi M, Lednicky JA, Glenn Morris J. 2 August 2019, posting date Mayaro as a Caribbean traveler: evidence for multiple introductions and transmission of the virus into Haiti. Int J Infect Dis doi: 10.1016/j.ijid.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White SK, Mavian C, Salemi M, Morris JG, Elbadry MA, Okech BA, Lednicky JA, Dunford JC. 2018. A new “American” subgroup of African-lineage Chikungunya virus detected in and isolated from mosquitoes collected in Haiti, 2016. PLoS One 13:e0196857. doi: 10.1371/journal.pone.0196857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florida Department of Health. 2019. Florida Arbovirus Surveillance: Week 52, December 22–28, 2019. http://www.floridahealth.gov/diseases-and-conditions/mosquito-borne-diseases/_documents/2019-week-52-arbovirus-surveillance-report.pdf.
- 22.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Boogaart GK, Tolosana-Delgado R, Bren M. 2018. compositions: compositional data analysis. R package version 1.40–2. https://CRAN.R-project.org/package=compositions.
- 28.Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. 2013. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One 8:e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes AD, Reid JN, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB. 2014. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tcherepanov V, Ehlers A, Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickett BE, Greer DS, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen CN, Jen W, Klem EB, Scheuermann RH. 2012. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses 4:3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32:1933–1942. doi: 10.1093/bioinformatics/btw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 36.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut A, Lam TT, Carvalho LM, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/bf02101694. [DOI] [PubMed] [Google Scholar]
- 43.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. 2011. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 60:150–160. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu G, Smith DK, Zhu H, Guan Y, Lam TT. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 46.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 47.Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 48.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinway SN, Dannenfelser R, Laucius CD, Hayes JE, Nayak S. 2010. JCoDA: a tool for detecting evolutionary selection. BMC Bioinformatics 11:284. doi: 10.1186/1471-2105-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 52.Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL. 1 March 2018, posting date Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNASeq read-proportion analyses. To analyze the read abundance of the RNASeq assay conducted on the mosquito samples from Manatee County, the ratio of DENV4-mapped reads to the total number of reads mapped for each site and year pool was calculated. The number of reads that mapped specifically to DENV4 was divided by the total number of reads mapped, the resultant values being shown above the bars on the graph. Proportion values are shown on the y axis of the graph. Download FIG S1, PDF file, 0.6 MB (598.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Assessment of phylogenetic quality for DENV4 strains. (a and c) Evaluation of the presence of phylogenetic signal satisfying resolved phylogenetic relationships among sequences was performed by likelihood mapping (IQ-TREE: http://www.iqtree.org/), which estimates the likelihood of each of the three possible tree topologies for each group of four sequences (quartet) in the data set using the best-fit nucleotide substitution model chosen according to the Bayesian information criterion (BIC). Quartets were considered resolved when the three likelihoods were significantly different (phylogenetic signal) and were considered unresolved, or partially resolved, when two likelihood values or all three likelihood values were not significantly different (phylogenetic noise). Percentages shown within each triangle indicate the proportion of resolved quartets (in the three corner areas) and the proportion of partially resolved (side areas) or unresolved (center) quartets. Extensive simulation studies have shown that side/center areas, including <40% of the unresolved quartets, can be considered robust in terms of phylogenetic signal (K. Strimmer, A. von Haeseler, Proc Natl Acad Sci U S A 94:6815–6819, 1997, https://doi.org/10.1073/pnas.94.13.6815; K. Strimmer, A. von Haeseler, Mol Biol Evol 13:964–969, 1996, https://doi.org/10.1093/oxfordjournals.molbev.a025664). (b and d) Substitution saturation, which decreases the phylogenetic information contained in the sequences, was assessed using DAMBE7 (http://dambe.bio.uottawa.ca/DAMBE/dambe.aspx) by plotting pairwise nucleotide (blue) transition (s) and (green) transversion (v) substitutions (y axis) versus pairwise genetic distances (x axis) determined with the Tamura and Nei 1993 (TN93) nucleotide substitution model (K. Tamura, M. Nei, Mol Biol Evol 10:512-526, 1993, https://doi.org/10.1093/oxfordjournals.molbev.a040023). Download FIG S2, PDF file, 0.8 MB (857.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Molecular clock and demographic prior model testing for phylogenetic analyses. Download Table S1, PDF file, 0.6 MB (663.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Assessment of temporal signal for DENV4 strains. The plot represents regression analysis of root-to-tip genetic distances assessed using TempEst v1.5. The positive slope (R2 = 0.7135) indicates the presence of temporal signal for the dataset. Download FIG S3, PDF file, 0.5 MB (502.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Amino acid differences between Manatee County, Haitian, and Senegal DENV4 genomes. Download Table S2, PDF file, 0.6 MB (631.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DENV4 3′ UTR analyses. (a) Alignment with the 3′ UTR of Manatee County DENV4. The 3′ UTR RNA sequence alignment is of DENV4 from Manatee County, DENV4 Haiti 2014 no. 2 (GenBank accession no. KT276273.1), and DENV4 Philippines H241 (GenBank accession no. KR011349.2) genomes. The DENV4 RNA sequence alignment was generated with CLC Sequence Viewer 8.0 (http://resources.qiagenbioinformatics.com/manuals/clcsequenceviewer/current/index.php?manual=Introduction_CLC_Sequence_Viewer.html). Some conserved DENV4 3′ UTRs are designated in black boxes in the figure, including repeated conserved sequence 2 (RCS2), conserved sequence 1 (CS1), conserved sequence 2 (CS2), and a 3′ upstream AUG region (3′ UAR). Different nucleotides are designated with different colors. (b) A diagram of the secondary structure of the DENV4 Manatee County 3′ UTR is depicted with key nucleotides and mutations highlighted and drawn in orange, correlating to node A or node B from Fig. 4B. Key secondary structure regions of the 3′ UTR are shown in black text as follows: dumbbell 1 (DB1) and DB2, pseudoknots 1 to 5 (PK1 to PK5), flavivirus nuclease-resistant RNA (fNR2), and the 3′ stem loop (3′ SL). Download FIG S4, PDF file, 0.8 MB (785KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Viral RNASeq read data are available in the NCBI Sequence Read Archive and Biosample archive under BioProject PRJNA547758. Genome sequence data for the Manatee County sequence are available in NCBI’s GenBank database (GenBank accession no. MN192436), and reference sequences are available in the GenBank database under the accession numbers provided in the text. Multiple coding DENV4 sequence alignments from the dN/dS analyses and alignments for the RNA secondary structure model in Fig. S4 in the supplemental material are available with relevant accession numbers in a Mendeley data set (https://data.mendeley.com/datasets/kwszjp63rb/draft?a=e11f9b80-bcfb-443b-918d-3016032ef3bd). The GenBank accession numbers for the sequences compared in the alignments shown in Fig. 5c and d are (from top to bottom in panel c and d, excluding AH011951.2 in panel d for which sequencing data of the strain is not available in the compared region of the genome) MN192436, AH011951.2, JF262782.1, KP140942.1, KT276273.1, MK514144.1, and MF004387.1.