Figure 6.
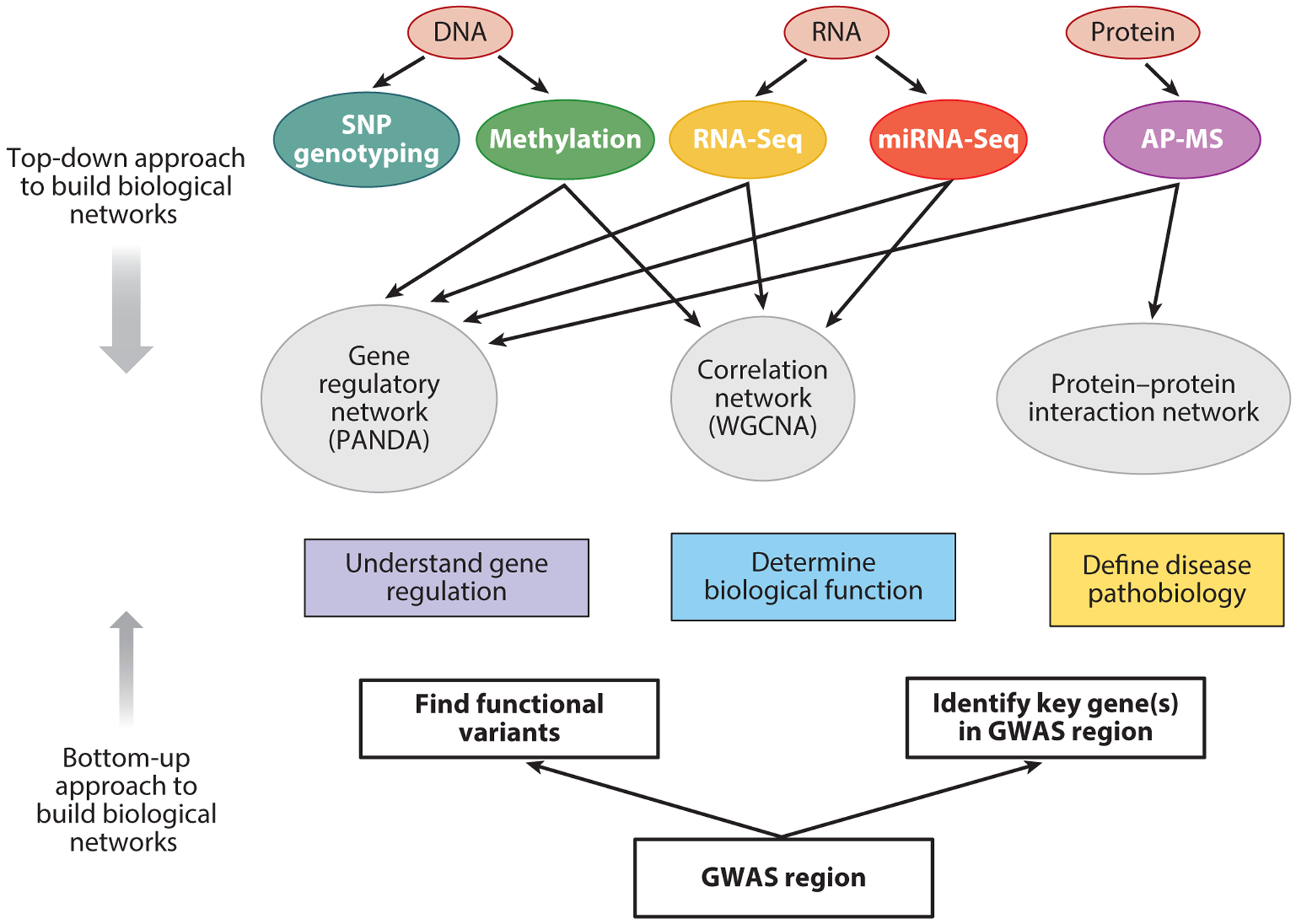
Comparison of top-down and bottom-up approaches to build biological networks. Biological networks can be built from the bottom up, starting with GWAS regions and identifying the key genes and the functional variants that impact those genes. Networks can be extended from those genes using tools that assess binding with other proteins (e.g., tandem affinity purification, coimmunoprecipitation) as well as hypothesis-based molecular biology experiments. Top-down approaches begin with Big Data assessments of key biological molecules such as DNA, RNA, proteins, and metabolites. Various types of networks can be built, including correlation-based networks, gene regulatory networks, and protein–protein interaction networks. Ultimately, bottom-up and top-down approaches may converge to give insights into gene regulation, biological function, and disease pathobiology relevant to COPD. The top-down approach is encompassed by the omics data and network methods shown above the rectangles labeled “Understand gene regulation,” “Determine biological function,” and “Define disease pathobiology,” whereas the bottom-up approach includes the components below those rectangles. Abbreviations: AP-MS, affinity purification–mass spectrometry; GWAS, genome-wide association study; SNP, single nucleotide polymorphism.
