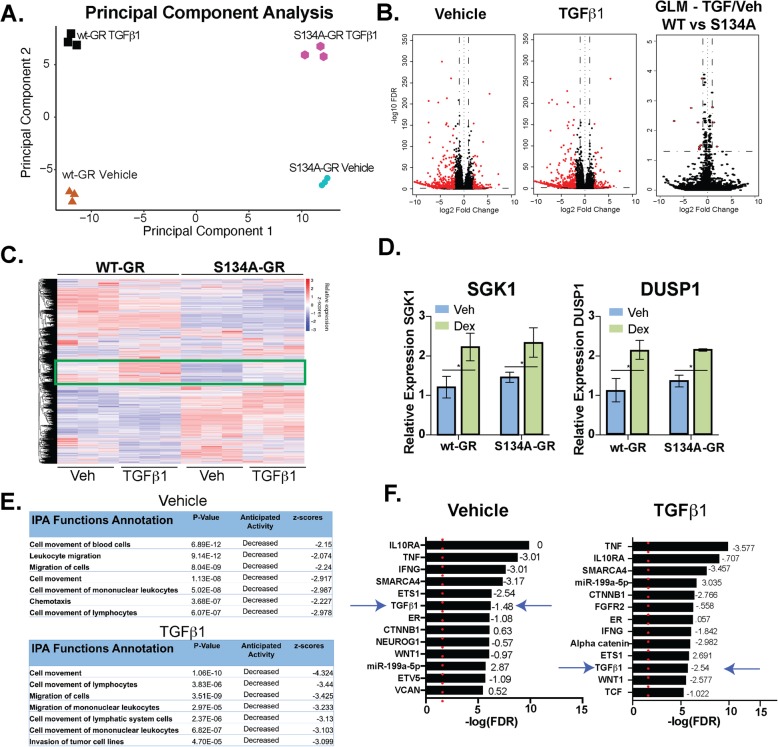
Fig. 5.
WT vs. S134A-GR transcriptomes in TGFβ1-treated MDA-MB-231 cells. a Principal component analysis plot derived from RNA-seq shows the differences in transcriptomes between MDA-MB-231 cells expressing either WT or S134A-GR cells and treated with vehicle or 10 ng/mL TGFβ1 for 6 h. b Volcano plot showing differential expression of genes in wt-GR+ compared to S134A-GR+ TNBC cells treated for 6 h with either Vehicle (left) or 10 ng/mL of TGFβ1 (middle). Volcano plot using the generalized linear model approach in EdgeR (comparisons of wt-GR and S134A-GR in response to TGF) to illustrate the effects of TGFβ1 treatment of cells expressing wt-GR relative to cells expressing S134A-GR (right). Red dots indicate genes with an absolute log2 fold-change of 1.0 or more and a false discovery rate of less than 0.05. Genes for GLM are detailed in Supplementary Table 1A. c Supervised heatmap shows significant differences in gene expression as measured by RNA-seq in TGFβ1-treated MDA-MB-231 cells expressing either wt-GR or S134A-GR. Rectangle indicates a cluster of interest in which genes are upregulated in response to TGFβ1 in wt-GR cells but not in the S134A-GR cells. All genes that were used for differential expression analysis are represented in this heatmap (n = 19,760). dDUSP1 and SGK1 mRNA levels were assessed using qRT-PCR following normalization to 18S expression. Mean expression of three independent replicates ± SD is shown. e Cell function and mechanism analyses of pathways via IPA of differentially expressed genes with an absolute log fold-change of 1.5 or more and a p-adjusted value of less than 0.05 in wt-GR relative to S134A-GR TNBC cells treated for 6 h with either Vehicle or TGFβ1 (10 ng/mL). f Upstream regulator analyses via IPA of differentially expressed genes with an absolute log fold-change of 1.5 or more and a p-adjusted value of less than 0.05 in wt-GR relative to S134A-GR TNBC cells treated with either Vehicle or 10 ng/mL of TGFβ1. Activation/inhibition z-scores are shown as well. Red dotted line indicates the significance value of 1.3 (p < .05)