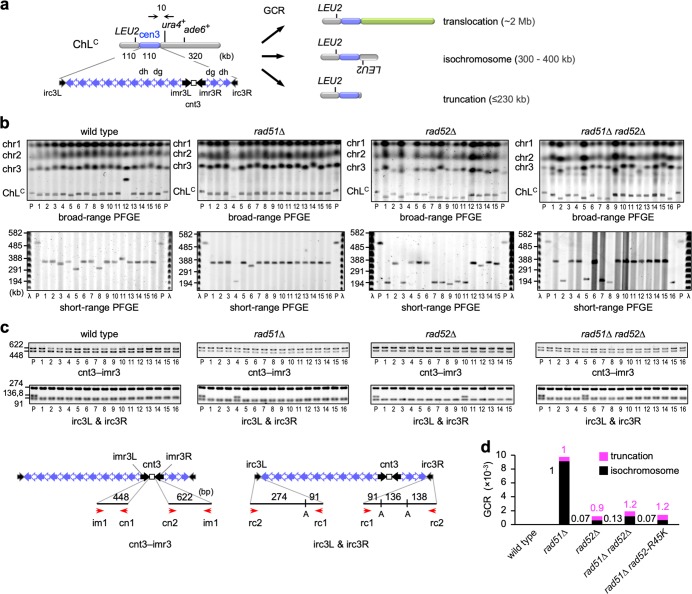
Fig. 2. Rad52 is required for isochromosome formation but not truncation.
a Three types of GCRs (translocation, isochromosome formation, and truncation) can be detected using ChLC. The three can be differentiated by length. b Chromosomal DNAs from parental strains (P) and independent GCR clones of wild type, rad51∆, rad52∆, and rad52∆ rad51∆ were separated by broad- and short-range PFGE and stained with EtBr (top and bottom rows, respectively). Positions of chr1, chr2, chr3, and ChLC (5.7, 4.6, ~3.5, and 0.5 Mb, respectively) in the parental strains are indicated on the left side of the broad-range PFGE panels (top row). Numbers on the left of the short-range PFGE panels (bottom row) indicate sizes of λ ladders (Promega). c PCR analysis of GCR products recovered from agarose gels. Both sides of cnt3–imr3 junctions were amplified and resolved by standard agarose gel electrophoresis (cnt3–imr3). Amplified irc3R and irc3L regions were digested with ApoI (irc3L & irc3R) prior to the electrophoresis. Positions of primers (red arrows) and ApoI sites are indicated at the bottom. A, ApoI. d Rates of truncation (magenta) and isochromosome formation (black). Rates relative to that of the rad51∆ strain are indicated. Uncropped images of the gels presented in b, c are shown in Supplementary Fig. 7. Source data for the graph in d are available in Supplementary Data 1.