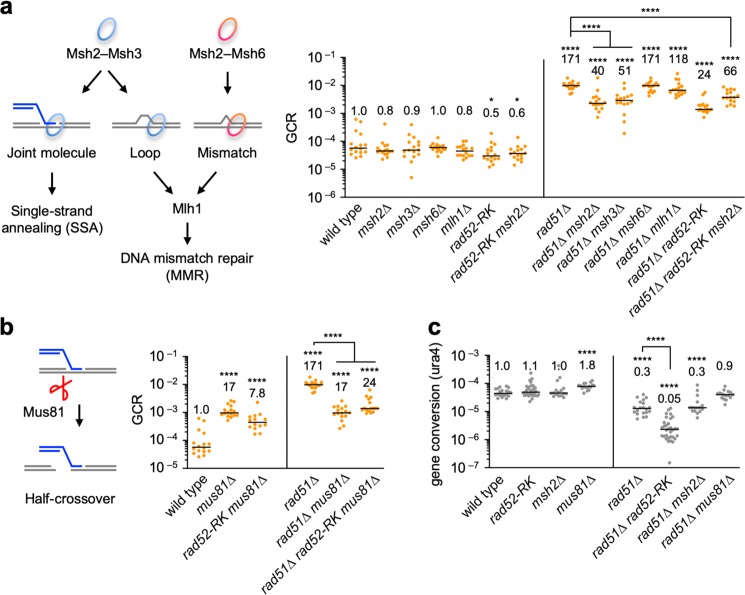
Fig. 5. Msh2, Msh3, and Mus81 are involved in the Rad52-dependent GCR pathway.
a GCR rates of the wild-type, msh2∆, msh3∆, msh6∆, mlh1∆, rad52-R45K, rad52-R45K msh2∆, rad51∆, rad51∆ msh2∆, rad51∆ msh3∆, rad51∆ msh6∆, rad51∆ mlh1∆, rad51∆ rad52-R45K, and rad51∆ rad52-R45K msh2∆ strains (TNF5369, 6618, 6867, 6869, 6620, 6627, 6599, 5411, 6649, 7081, 6908, 6651, 6616, and 6697, respectively). The roles of Msh2–Msh3, Msh2–Msh6, and Mlh1 in single-strand annealing (SSA) and DNA mismatch repair (MMR) are illustrated on the left side of the graph. b GCR rates of the wild-type, mus81∆, rad52-R45K mus81∆, rad51∆, rad51∆ mus81∆, and rad51∆ rad52-R45K mus81∆ strains (TNF5369, 5669, 6614, 5411, 5974, and 6648, respectively). The role of Mus81 endonuclease in half-crossover formation is illustrated on the left of the graph. c Gene conversion rates at the ura4 locus in the wild-type, rad52-R45K, msh2∆, mus81∆, rad51∆, rad51∆ rad52-R45K, rad51∆ msh2∆, and rad51∆ mus81∆ strains (TNF3631, 5995, 6128, 6518, 3635, 6021, 6136, and 6569, respectively). *P < 0.05; ****P < 0.0001. Source data for the graphs in a–c are available in Supplementary Data 1.