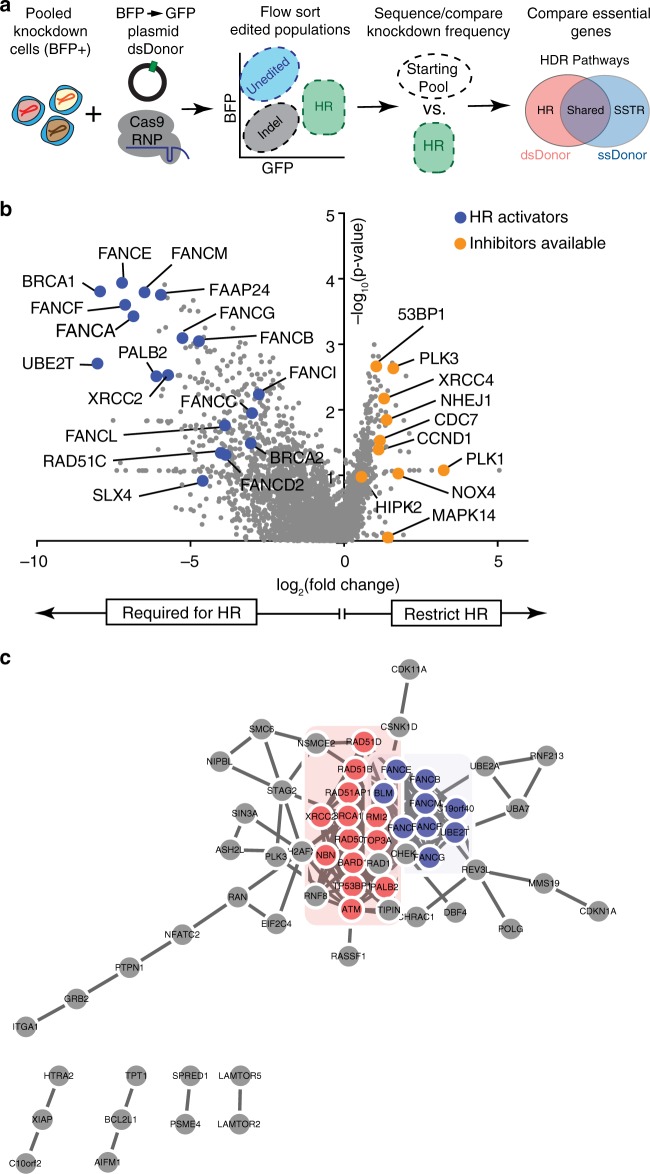
Fig. 1. A pooled CRISPR screen reveals pathways that regulate templated repair using Cas9-RNP and a plasmid dsDonor.
a Schematic showing BFP → GFP CRISPRi screening strategy. Pooled K562-CRISPRi cells that stably express BFP and a library of gRNAs targeting DNA metabolism genes are further edited with Cas9-RNP that cuts within BFP and a plasmid dsDonor template that contains a promoterless copy of GFP. Cas9 gene editing results in three populations: Unedited (BFP+GFP−), Indel (BFP−GFP−), and HR (BFP−GFP+). Guide RNA frequencies in the HR population were quantified and compared to the unsorted population to identify genes that promote/restrict HR. These genes were compared to data from a prior screen using ssDNA donor8. b Genes regulating dsDonor HR. A volcano plot of pooled screen hits showing the Fanconi Anemia pathway (blue) and HR repressors with available small molecule inhibitors (orange). Data presented from n = 2 screen replicates. c Genetic regulators of HR participate in physical interactions with one another. Significant (p < 0.005, Mann–Whitney test) hits that impacted HR were cross-referenced with the STRING protein interaction database12. Factors involved in homologous recombination (HR, red) or Fanconi Anemia (FA, blue) pathways were identified by GO term. Only regulators appearing in the screen and having at least one interaction with another screening hit are shown.