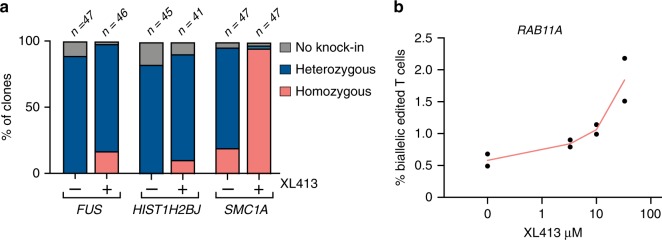
Fig. 5. XL413 increases biallelic editing at endogenous loci.
a XL413 increases homozygous editing in K562 cells. Cells were nucleofected with RNPs targeting three different genomic loci (FUS, HIST1H2BJ, and SMC1A) and plasmid dsDonor to knock a GFP reporter protein into the C-terminal end of the gene. Nucleofected cells were untreated or treated with 10 μM XL413 for 24 h. GFP+ cells were single-cell sorted 4 days post nucleofection, clonally expanded, and allelic editing frequencies quantified by junction PCR (Supplementary Fig. 5a, b). The total number of clones analyzed is noted above each column with the proportion of heterozygous, homozygous, or no knock-in (GFP+ but no detection of GFP sequence at on-target site) genotype shown per condition. b XL413 increases biallelic editing in T cells. CD3+ T cells were nucleofected with RNPs targeting the RAB11A locus and two linear dsDonors encoding N-terminal fusions of either mCherry or BFP and treated with the indicated concentrations of XL413 for 24 h. Single-fluorescent or double-fluorescent reporter populations were quantified by flow cytometry (Supplementary Fig. 5c, d). Shown is the frequency of biallelic dual-positive integration (BFP+/mCherry+) events displayed as mean ± SD (n = 2 biological replicates). Source data are available in the Source Data file.