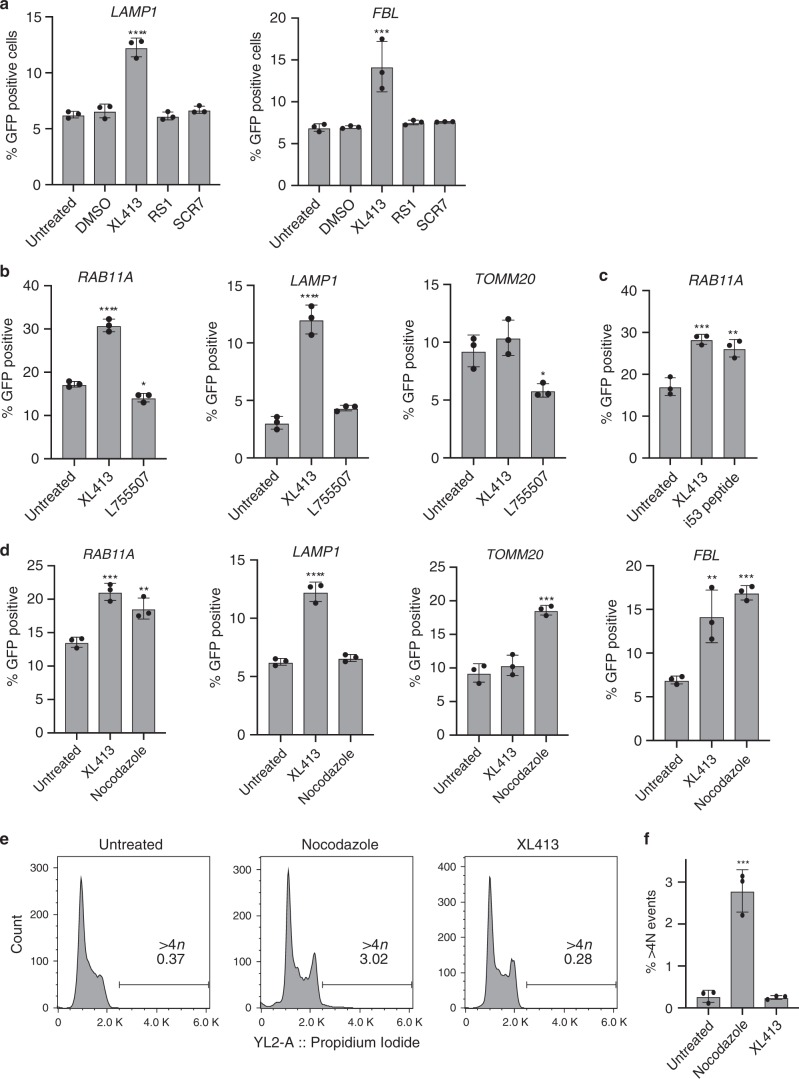
Fig. 6. Comparisons between XL413 and other small molecule HDR-boosting strategies in K562 cells.
a Direct comparison between XL413 and RS1 or SCR7. Cells were nucleofected with RNP targeting the LAMP1 or FBL loci with dsDonors encoding C-terminal fusions to GFP, cultured in XL413 (33 μM), SCR7 (1 μM) or RS-1 (10 μM) for 24 h, and editing rates were measured 4 days post nucleofection by flow cytometry. b Comparison between XL413 and L755507. Cells were nucleofected with RNPs targeting the RAB11A, LAMP1, or TOMM20 loci with dsDonors encoding N-terminal (RAB11A) or C-terminal (LAMP1, TOMM20) fusions to GFP, cultured in XL413 (10 μM) or L755507 (5 μM) for 24 h, and editing rates were measured 4 days post nucleofection by flow cytometry. c Comparison between XL413 and i53. Cells were nucleofected with RNP targeting the RAB11A locus with dsDonor encoding an N-terminal fusion to GFP. i53-treated samples were lipofected with (3 μg) plasmid encoding i53 and BFP 24 h prior to nucleofection with editing reagents. XL413-treated samples were cultured in 10 μM XL413 for 24 h after nucleofection, then editing outcomes were measured 4 days post-nucleofection by flow cytometry. i53-transfected cells were gated using a BFP fluorescent marker expressed from the i53 expression plasmid. d Direct comparison between XL413 and nocodazole. Cells were nucleofected with RNP targeting the RAB11A, LAMP1, TOMM20, or FBL loci with dsDonors encoding N-terminal (RAB11A) or C-terminal (LAMP1, TOMM20, and FBL) fusions to GFP. Samples were treated with nocodazole (33 ng mL−1) for 24 h before nucleofection. XL413-treated samples were cultured in 10 μM XL413 for 24 h after nucleofection. Editing rates were measured 4 days post-nucleofection by flow cytometry. e 1Cells were treated with nocodazole or XL413 for 24 h and then released into normal media for four days prior to propidium iodide staining and flow cytometry. Example plots are shown for untreated, nocodazole, and XL413 treated samples. f Quantification of frequency of cells with a > 4 N DNA content (indicative of failed mitoses) from e. All values are shown as mean ± SD (n = 3 biological replicates). Statistical significances were calculated by ordinary one-way ANOVA and Dunnet’s multiple comparison test (adjusted p-values are reported as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Source data are available in the Source Data file.