Figure 1.
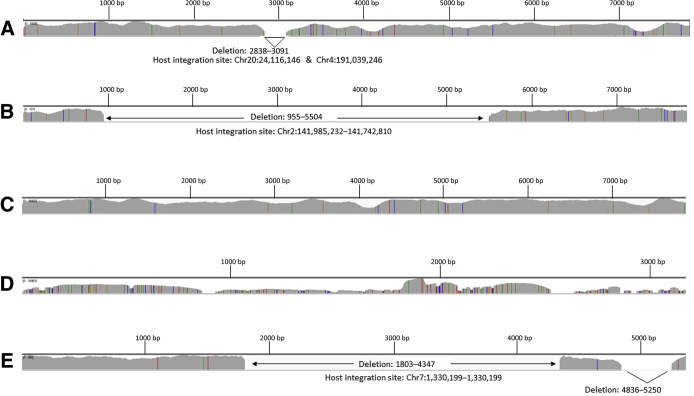
Virus target coverage obtained with ViroPanel. Integrated Genome Viewer (IGV) coverage plots of selected ViroPanel samples. A: ViroPanel detected 5581 viral reads/million to support presence of human papillomavirus (HPV) 16 in sample 28 (previously tested HPV negative via in situ hybridization) occurring with a 253-bp deletion (HPV16: 2838 to 3091 bp) and evidence of viral integration. B: Sample 7 shows evidence of HPV18 integration and a 4549-bp deletion (HPV18: 955 to 5504 bp), identical integration features observed in a second biopsy taken from the same patient on a later date (sample 9). C: ViroPanel detected 94,340 viral reads/million supporting HPV16 infection in sample 16, which was previously tested and designated as HPV45 because of a G > A mutation at position HPV16: 7008 that eliminated a Pst1 restriction enzyme site used for PCR–restriction fragment length polymorphism analysis (see Discrepant Sample Review and Sensitivity and Specificity). D: Hepatitis B virus coverage observed in sample 21. E: Merkel cell polyomavirus coverage observed in Merkel cell carcinoma sample 80, with evidence of viral integration; viral and host break points observed in matching patient-derived xenograft, sample 95. Genomic start positions in IGV plots correspond to first nucleotide in each of the virus genomic references (Table 1). Chr, chromosome.