Figure 2. Preferential Effects of PRMT5 or Type I PRMT Inhibition on SF-Mutant AML over WT Counterparts In Vitro.
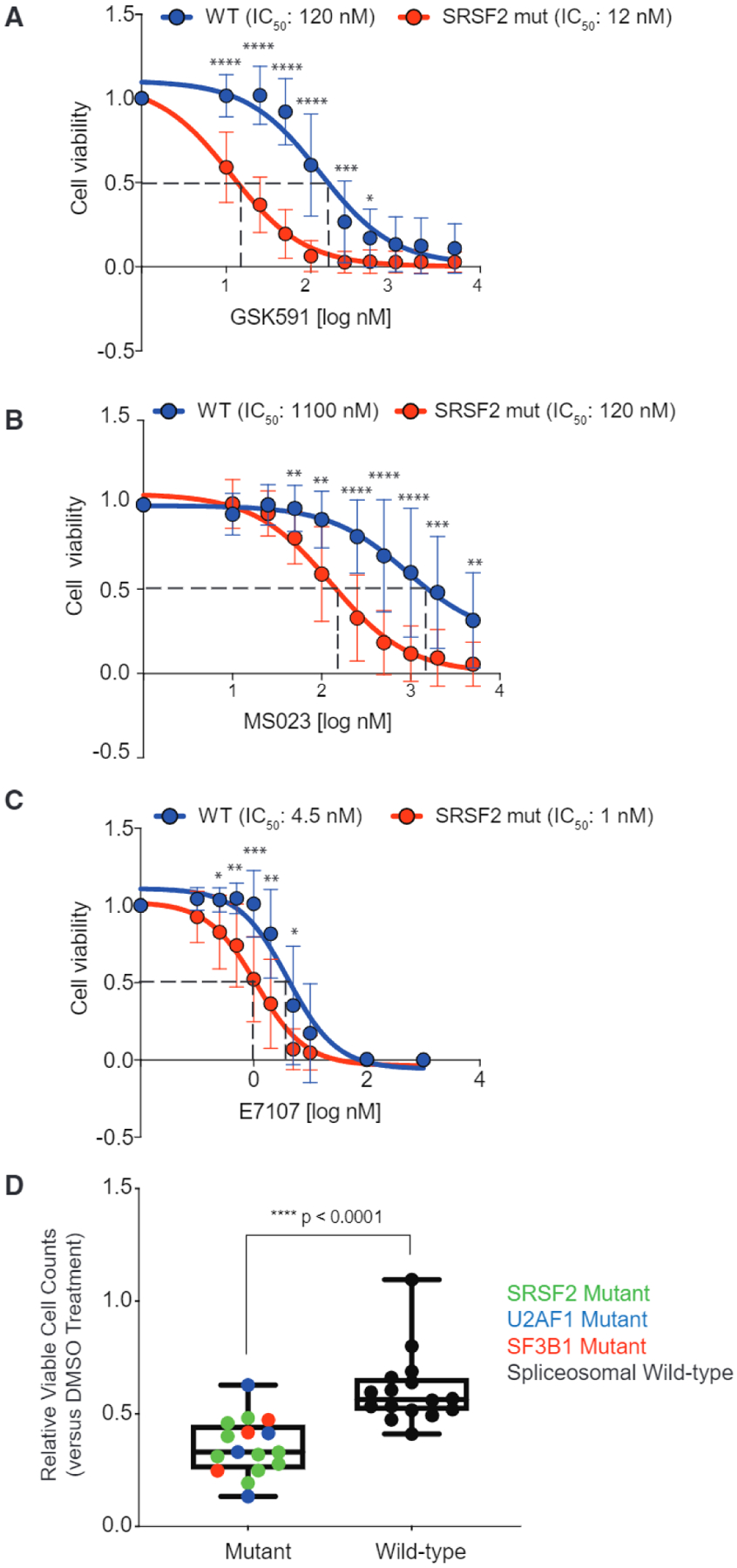
(A–C) Relative cell viability of (MLL-AF9/Vav-cre Srsf2WT and MLL-AF9/Vav-cre Srsf2P95H treated with GSK591 (A), MS023 (B), or E7107 (C), and normalized to control (SGC2096a for PRMT5, MS094 for PRMT1, and DMSO for E7107). Samples were prepared in four to six replicates and averages were calculated, error bars represent SD. Student’s t test was used for statistical analysis.
(D) Relative viable cell counts of AML patient samples to GSK591 based on spliceosomal gene mutation status. Primary AML cells with SF-mutations (n = 16) or WT for SRSF2, U2AF1, and SF3B1 (n = 16) were incubated with DMSO or GSK591 (0.5 mM) for 6 days. Cells were subjected to flow cytometry to detect 7-AAD-negative, YO-PRO1-negative, viable cells. Relative viable cell numbers were compared with Welch’s t test. Boxplot top line of whisker denotes the highest value in dataset and bottom line of whisker denotes the lowest value in dataset, box spans interquartile range and line in box indicates median.
*p = 0.01–0.05, **p = 0.001–0.01, ***P = 0.0001–0.001, ***p < 0.0001. See also Figure S1 and Table S2.
