Figure 6. Global Profiling of PRMT Substrates at Single-Site Resolution by Quantitative Liquid Chromatography-Tandem Mass Spectrometry.
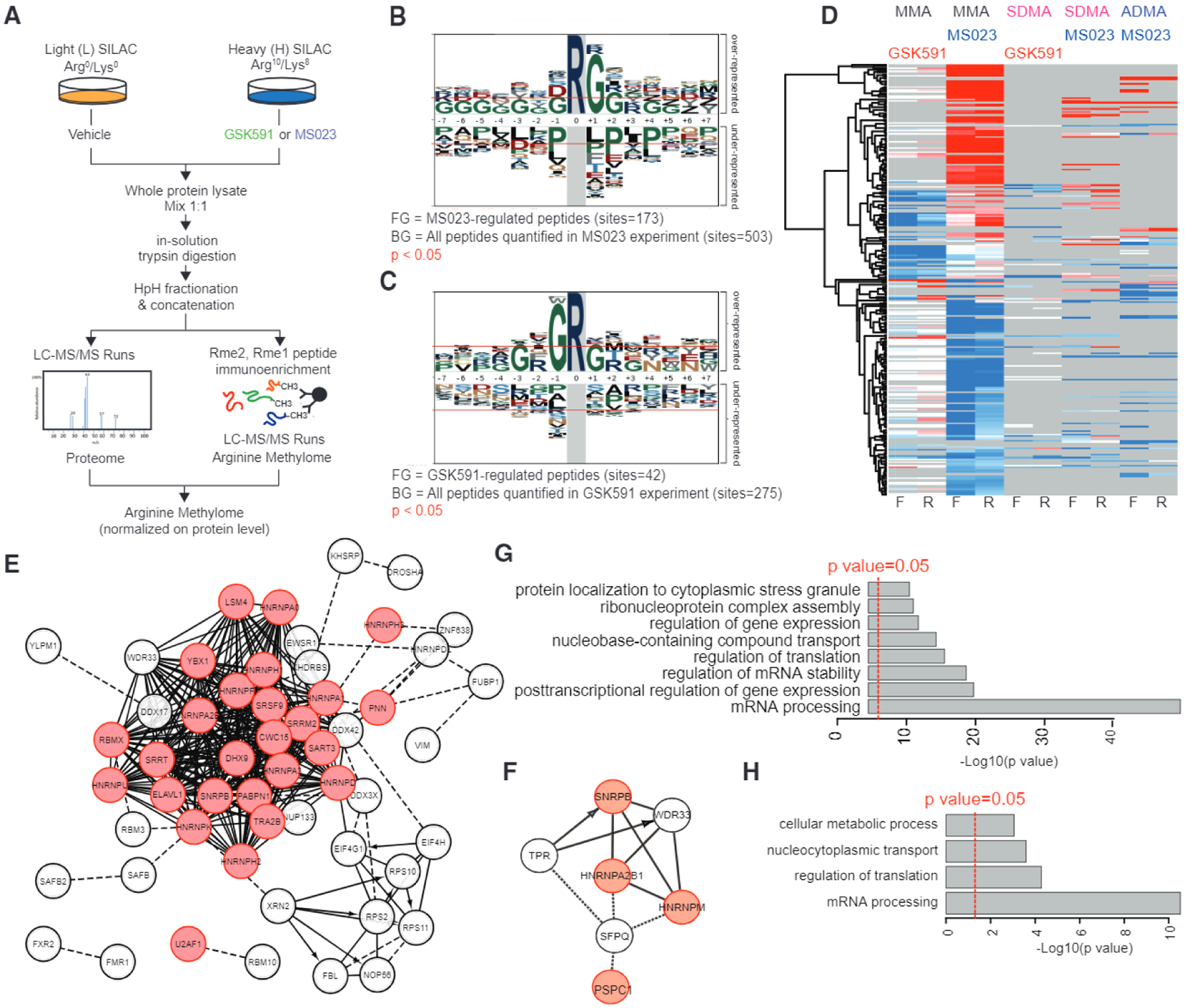
(A) Workflow of the SILAC methyl-R-proteomic experiments carried out to identify mono- and dimethylarginine substrates regulated by GSK591 and MS023 in NB4 leukemia cells.
(B and C) Sequence motif analysis indicates the consensus sequences significantly enriched in the methyl-peptides regulated by MS023 (B) or GSK591 (C).
(D) Heatmap of log2 SILAC ratios of each methyl-peptide identified and quantified from the SILAC experiment diagrammed in (A). For each pharmacological treatment two biological replicates, in forward and reverse SILAC mode, were carried out and different degrees of methylation were enriched and profiled. Only peptides reproducibly regulated in at least one pair of forward-reverse experiment are visualized in the heatmap.
(E and F) Network analysis of the proteins displaying methylation changes upon treatment with MS023 (E) or GSK591 (F). RNA-binding proteins are highlighted in red.
(G and H) Functional analysis of changing methylated proteins highlight the biological process terms significantly enriched among proteins displaying methylation changes upon treatment with MS023 (G) and GSK591 (H).
