Figure 6. Loss of Ect2 Results in Binucleation of CMs.
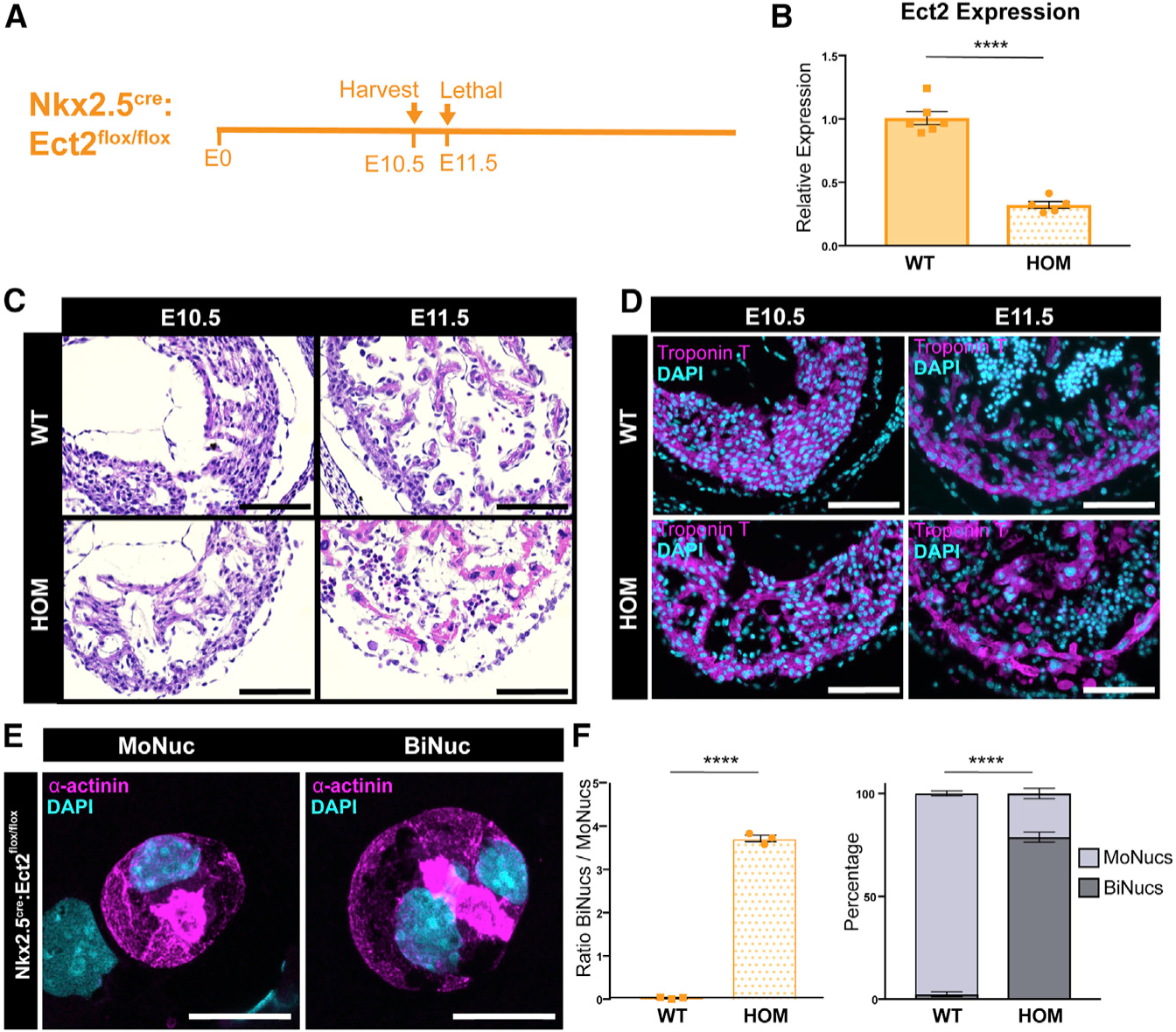
(A) Time course of Nkx2.5cre:Ect2flox/flox KO.
(B) Expression values of the Ect2 transcript in Nkx2.5cre:Ect2flox/flox (HOM; homozygous knockout) samples compared to Ect2flox/flox and Ect2flox/+ (wild-type [WT]) samples at E10.5 by qPCR. Data are reported as mean ± SEM. n = 5–6 animals per group.
(C and D) Representative tissue sections of WT and HOM hearts at E10.5 and E11.5 stained with H&E (C) or an antibody against cardiac troponin T (D) reveals that the Nkx2.5cre:Ect2flox/flox allele is lethal by E11.5. Scale bar, 100 μm. (D) Representative images of a MoNuc and BiNuc present in E10.5 HOM samples. Single-cell suspensions were cytospun and stained with an antibody against α-actinin. Scale bar, 20 μm.
(E) Quantification of the ratio of BiNucs to MoNucs and percentages of MoNucs and BiNucs in E10.5 WT and HOM samples. Data are reported as mean ± SEM. n = 3 animals per group. p values are determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
