Fig. 1. SLC7A11 promotes the PPP flux.
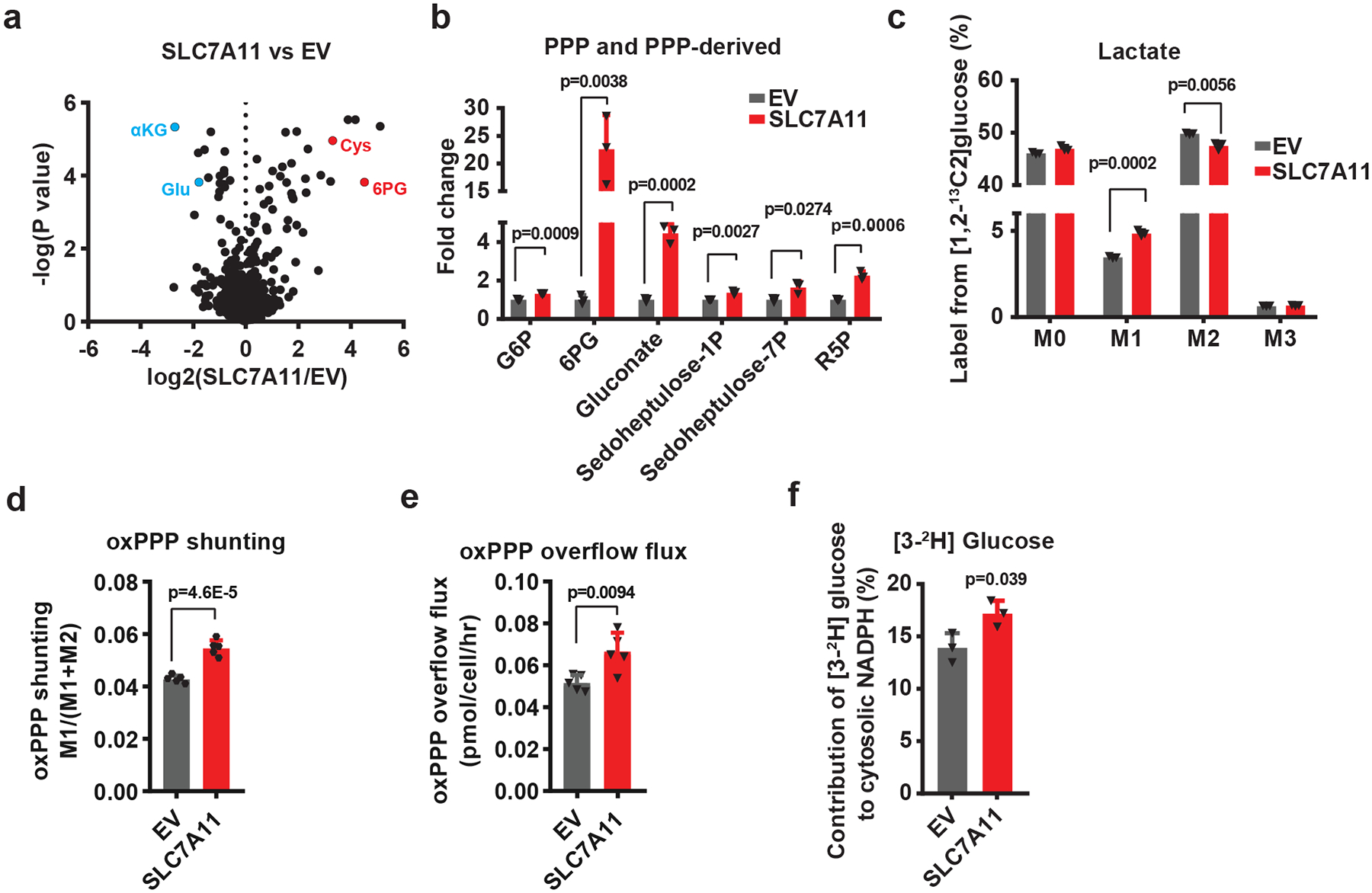
a, Volcano plot showing metabolomic profiling for SLC7A11-overexpressing 786-O cells compared with empty vector (EV) cells. n=3 independent experiments. b, Bar graph showing the fold changes of PPP and PPP-derived intermediates induced by SLC7A11 overexpression in 786-O cells. n=3 independent experiments. c, Mass isotopomer distribution of lactate produced from [1,2-13C2]-glucose in EV and SLC7A11-overexpressing 786-O cells. n=3 independent experiments. d, Relative flux of glucose carbon into the oxidative pentose phosphate pathway (oxPPP) in EV and SLC7A11-overexpressing 786-O cells. n=5 independent experiments. e The overflow flux through oxidative PPP in EV and SLC7A11-overexpressing 786-O cells. n=5 independent experiments. f, Contribution of glucose-derived deuterium labelled NADPH (via oxPPP) to cytosolic NADPH in EV and SLC7A11-overexpressing 786-O cells. n=3 independent experiments. In (f), data are plotted as mean ±95% confidence interval (CI). Other error bars are mean ± s.d.; all p values were calculated using two-tailed unpaired Student’s t-test. Numeral data are provided in Statistics Source Data Fig. 1. Abbreviations: αKG, α-ketoglutarate; Glu, glutamate; Cys, cysteine; 6PG, 6-phospho-gluconate; PPP, pentose phosphate pathway; G6P, glucose-6-phosphate; G6PD, glucose-6-phosphate dehydrogenase; PGD, 6-phosphogluconate dehydrogenase; R5P, ribose-5-phosphate; F6P, fructose-6-phosphate; GA3P, glyceraldehyde-3-phosphate; PYR, pyruvate; Lac, lactate; M0–3, 0–3 13C atoms labeled metabolites.
