Fig. 6. Phylogenetic and biochemical analysis of CYP72A proteins from three Brassicaceae plants, rice, and soybean.
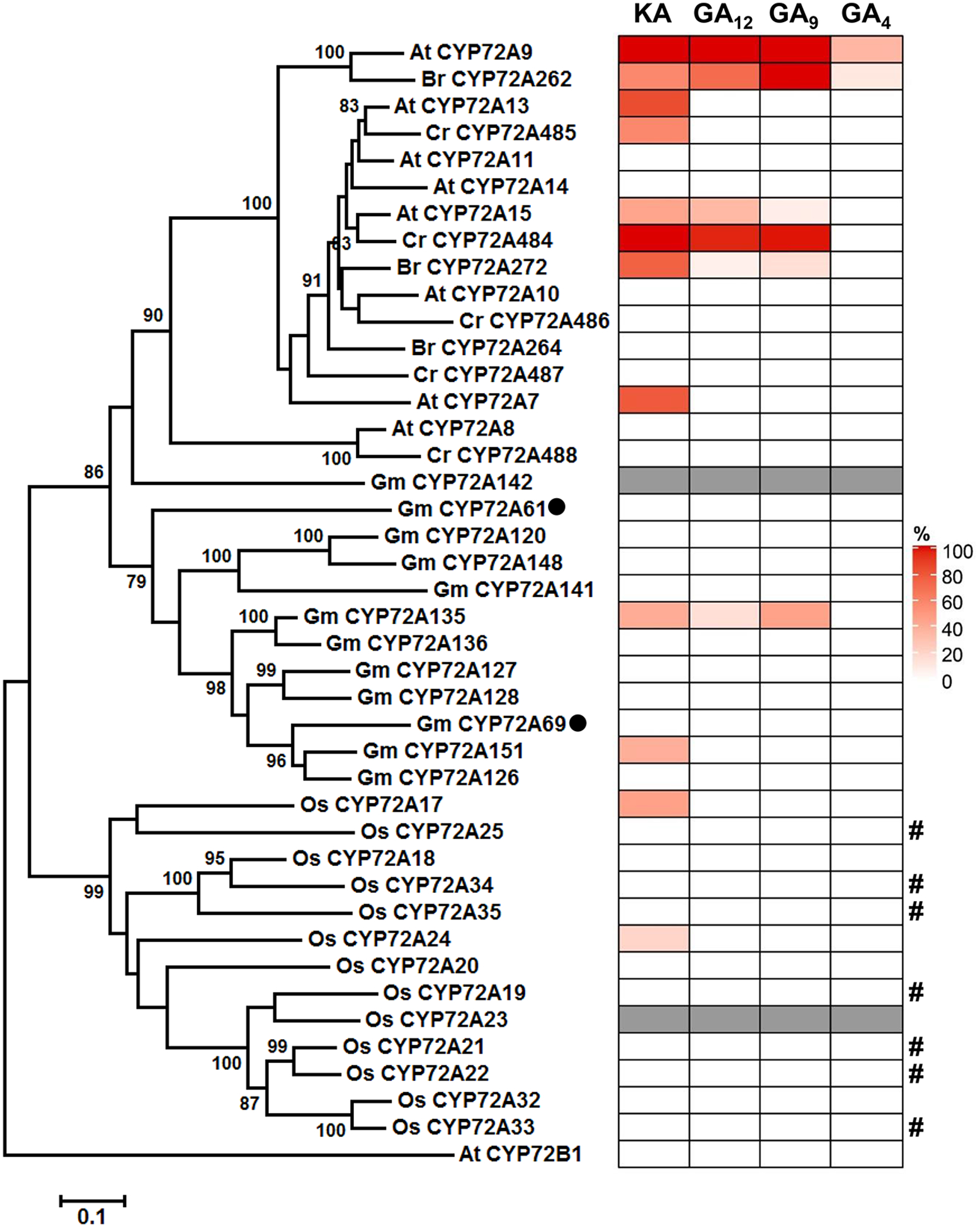
Gray color means no gene cloned in this study. GmCYP72A61 and GmCYP72A69, previously identified as triterpene oxidase, are indicated by black circles. Relative activity of various CYP72A proteins is expressed as the substrate conversion ratio (%). Values represent means from two independent experiments. It is noteworthy that the product of AtCYP72A15 and GmCYP72A135 using GA9 as the substrate is a mixture of GA20 and one unidentified hydroxylated GA9 (Supplementary Figs. 12 and 13). #, Inactivity with tested GA substrates might just reflect unsuccessful P450 protein expression in WAT11 yeast strain, which not detected by western blot (Supplementary Fig. 16).
