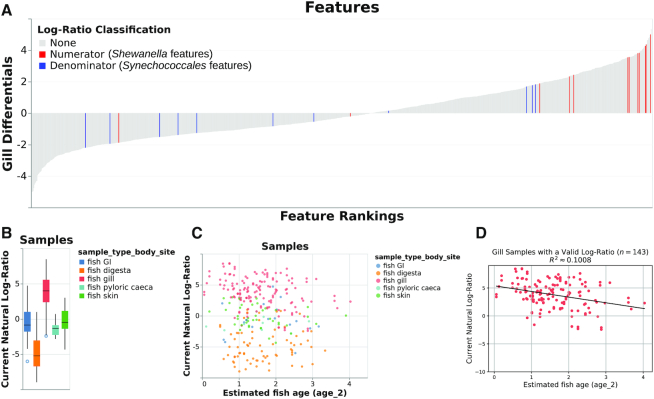
Figure 1.
Various outputs from the case study showing the log-ratio of classified Shewanella features to classified Synechococcales features. (A) ‘Rank plot’ showing differentials computed based on association with gill samples, using seawater samples as a reference category in the regression. The term ‘log-ratio classification’ only refers to the currently selected log-ratio in the Qurro visualization: in this case, this is the log-ratio of classified Shewanella features to classified Synechococcales features. To show the rankings of these ‘selected’ features relative to the remaining features in the dataset, these features are colored in the rank plot: Shewanella features are colored in red, and Synechococcales features are colored in blue. The remaining features, colored gray, have a ‘log-ratio classification’ of None because they are not involved in the selected Shewanella-to-Synechococcales log-ratio. (B) ‘Sample plot’ in boxplot mode, showing samples’ Shewanella-to-Synechococcales log-ratios by sample body site. Note that only 285 samples are represented in this plot; other samples were either filtered out upstream in the analysis or contained zeroes on at least one side of their log-ratio. (C) ‘Sample plot,’ showing a scatterplot of samples’ selected log-ratios versus estimated fish age. Individual samples are colored by body site. As in panel B, only 285 samples are present. (D) Ordinary-least-squares linear regression (R2 ≈ 0.1008) between estimated fish age and the selected log-ratio for just the 143 gill samples shown in B and C, computed outside of Qurro using scikit-learn (24) and pandas (12) and plotted using matplotlib (30).