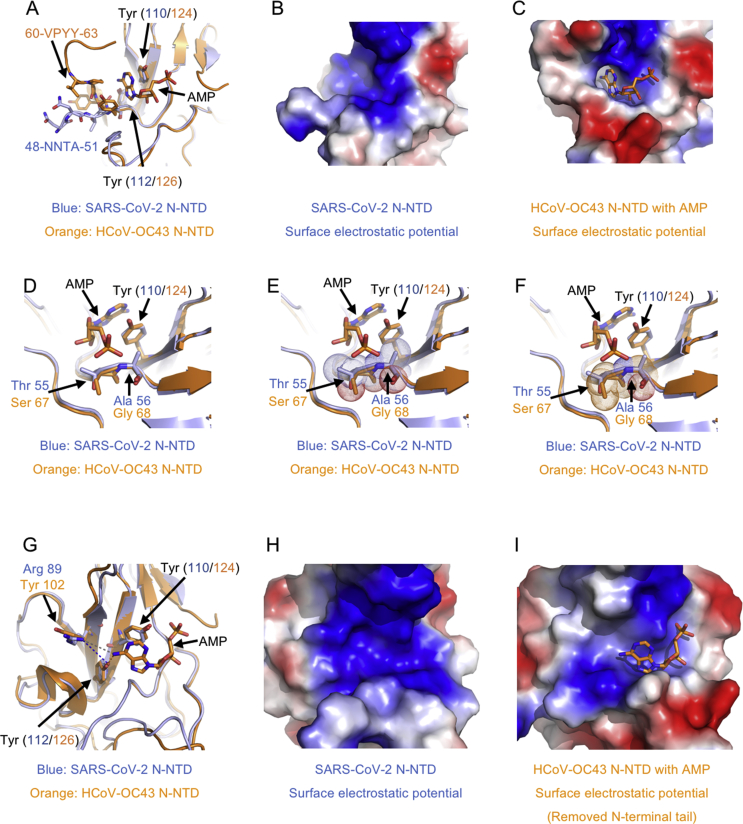
Figure 4.
A potential unique drug target pocket in SARS-CoV-2 N-NTD. (A) Detailed view of ribonucleotide binding pocket in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. AMP, interacting residues and equivalents are highlighted with stick representation. (B) Electrostatic surface of the potential ribonucleotide binding pocket on SARS-CoV-2 N-NTD. (C) Electrostatic surface of the ribonucleotide binding pocket on HCoV-OC43 N-NTD. (D) Detailed view of phosphate group binding site in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. (E) Dot representation of SARS-CoV-2 residues Thr55 and Ala56, which indicates potential steric clashes with the ribonucleotide phosphate group. (F) Dot representation of HCoV-OC43 N-NTD residues Ser67 and Gly68. (G) Detailed view of the nitrogenous base binding site in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. (H) Electrostatic surface of the potential ribonucleotide nitrogenous base binding pocket on SARS-CoV-2 N-NTD. (I) Electrostatic surface of the ribonucleotide nitrogenous base binding pocket on HCoV-OC43 N-NTD. In electrostatic surface potential panels, blue denotes positive charge potential, while red indicates negative charge potential. The potential distribution was calculated by Pymol. The values range from –6kT (red) to 0 (white) and to +6kT (blue), where k is the Boltzmann constant, and T is the temperature.