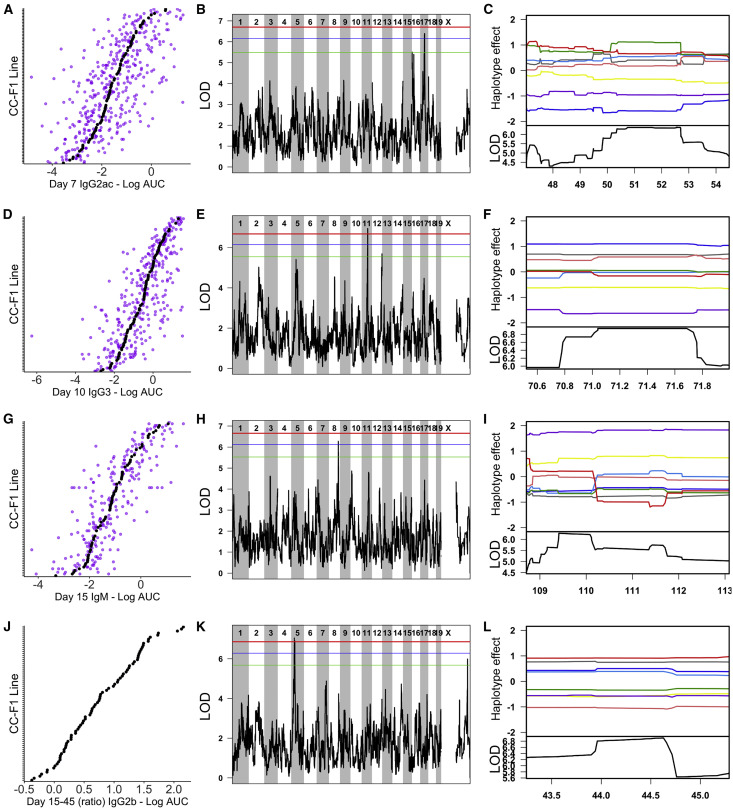
Figure 3.
Representative Ari QTLs Associated with Variation in IAV-Specific Antibody Responses
(A–C) show Ari1, D–F show Ari2, G–I show Ari3, and J–L show Ari4.
(A, D, G, and J) Phenotypic distributions for the traits mapped to Ari1–Ari4, respectively. Each plot is independently ordered by the CC-F1 means for that trait (black points) and also shows the individual mice (purple points). The exception is (J), which only shows mean values, as the ratios of antibody response between time points were calculated using the mean value for each F1 at the relevant individual time points.
(B, E, H, and K) show the associated QTL LOD plots (significance score across the genome) for Ari1–Ari4. Significance thresholds are shown in red (genome-wide p = 0.05), blue (p = 0.1), and green (p = 0.2). Following identification of QTLs, we determined the causal haplotypes driving these responses (C, F, I, and L). Each plot is zoomed in to the relevant QTL loci on the x axis. The lower black line shows the LOD score for that region, and the colored lines display the estimated effect of each of the 8 CC founder haplotypes (A/J = yellow, C57BL/6J = gray, 129S1/SvImJ = pink, NOD/ShiLtJ = dark blue, NZO/HlLtJ = light blue, CAST/EiJ = green, PWK/PhJ = red, and WSB/EiJ = purple). Causal haplotype groups are determined based on direction and distance from mean effect and the largest split distance between lines (e.g., in I, the WSB line is furthest away from all others).