Table 2.
CypD SAR of optimized ureas derived from merging of fragments 3 or 4 with the reference inhibitor 2.
| # | Structure | Biochemical CypD FP assay |
Biochemical PPIase assay IC50 (µM)c | SPR binding CypD |
||
|---|---|---|---|---|---|---|
| IC50 (µM)a | LEb | KD (µM)d | LEe | |||
| 14 | 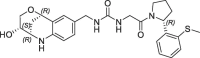 |
0.060 | 0.28 | 0.004 | 0.006 | 0.31 |
| 15 | 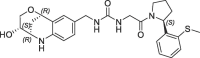 |
3.6 | 0.21 | nt | 0.630 | 0.23 |
| 16 | 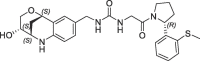 |
1.7 | 0.23 | nt | 0.670 | 0.23 |
| 17 | 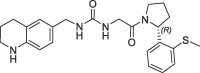 |
3.5 | 0.24 | 7.4 | 1.10 | 0.25 |
| 18 | 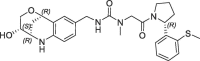 |
10 | 0.19 | nt | 7.0 | 0.19 |
| 19 | 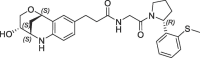 |
54 | 0.17 | nt | 39 | 0.17 |
| 20 | 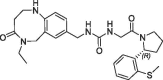 |
0.190 | 0.26 | nt | 0.180 | 0.25 |
| 21 | 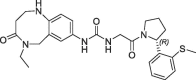 |
>100 | nt | 260 | 0.13 | |
| 22 |  |
4.1 | 0.21 | nt | 2.3 | 0.21 |
| 23 |  |
>100 | nt | 420 | 0.12 | |
a,bCypD biochemical FP and PPIase assays. c,eLigand binding efficiencies (LE) based on biochemical FP IC50s or SPR KDs. dCypD SPR binding assay. nt = not tested.
