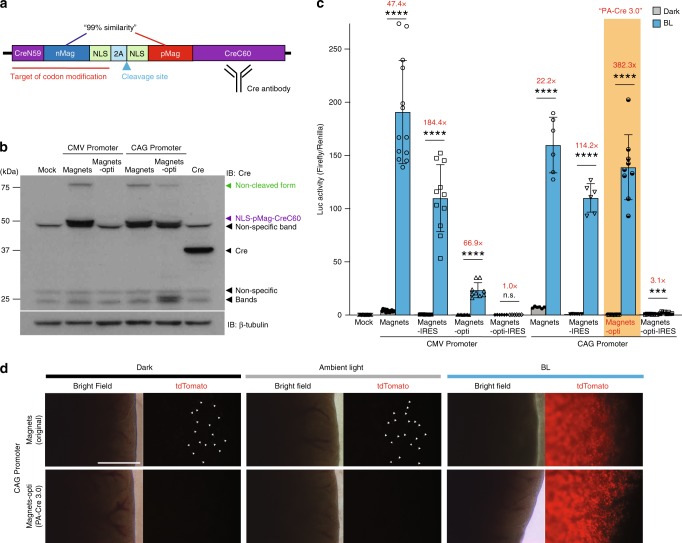
Fig. 2. Development and characterization of PA-Cre 3.0.
a Schematic representation of PA-Cre components indicating codon modification site, 2A cleavage site and Cre antibody recognition site. b Representative western blotting images to compare CMV and CAG promoters for testing Magnets and Magnets codon-optimized PA-Cre (Magnets-opti). All protein expressions were accomplished in the dark condition (single independent experiments). c Comparisons of the original Magnets- and Magnets-opti-based PA-Cre using CMV or CAG promoter with 2A or IRES sequences (Magnets-IRES, Magnets-opti-IRES) using dual Luc assay. Luc assays were conducted in HEK 293T with double-floxed inverted Fluc. The upper diagram shows an illumination protocol used for the assays (blue LED, 447.5 nm, 8.28 W/m2, repeated 20 s light and 60 s dark for 12 h) (n.s. not significant, ***P ≦ 0.0005; ****P < 0.0001; dark v.s. BL using Two-tailed t test, Biologically independent samples; Mock, CMV-Magnets: n = 13, CMV-Magnets-IRES n = 11, CMV-Magnets-opti n = 9, CMV-Magnets-opti-IRES, CAG-Magnets, CAG-Magnets-IRES: n = 6, CAG-Magnets-opti n = 9, CAG-Magnets-opti-IRES2 n = 8, mean ± s.d.). Red values show fold induction of Luc with BL compared with dark. d Comparison of CAG-driven Magnets-opti-based PA-Cre (PA-Cre 3.0) with the original version of PA-Cre (Magnets-based) using fluorescence imaging in mouse livers in vivo. Fluorescence liver imaging of tdTomato expression was conducted in Rosa26Ai14/WT (Ai14: Floxed-tdTomato heterozygote) live mice transiently injected with CAG-Magnets or CAG-Magnets-opti plasmids, respectively in the dark, ambient light and under blue-light illumination (470 ± 20 nm, 200 W/m2, 16 h continuous) (n = 2–6 mice/group, Scale bar: 1 mm). White arrowheads show tdTomato-positive cells in the livers in the dark and ambient light conditions. Source data are provided as a Source Data File.