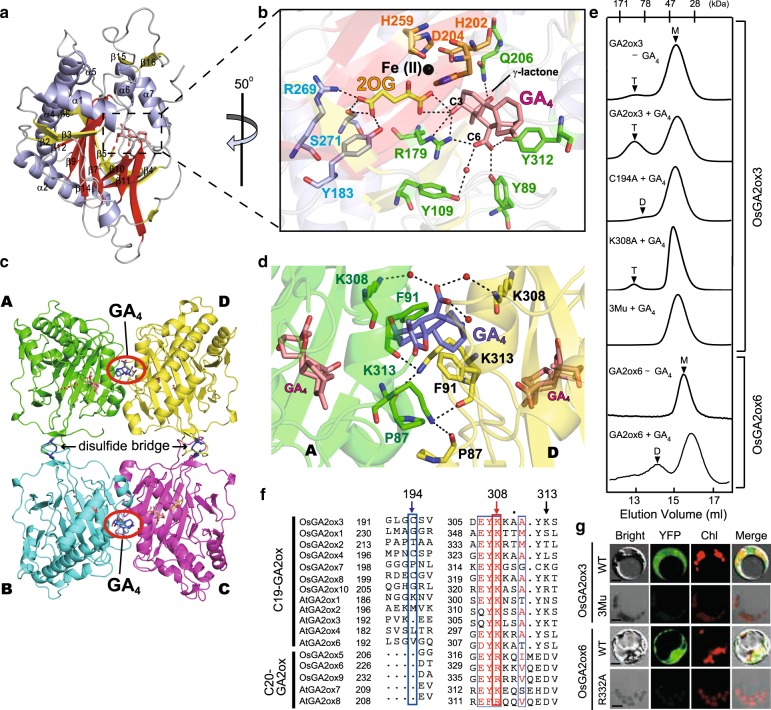
Fig. 1. OsGA2ox3 forms GA-mediated tetramer structure.
a Crystal structure of OsGA2ox3. The α-helices are shown in blue, the common core fold is shown in red and none core β-strands are in yellow. b Close-up view of the active site. Important active site residues 2OG and GA4 are shown in yellow and pink with stick form, respectively. Fe(II) is colored in black. The amino acids interacting with 2OG (R269, S271, and Y183 shown in blue characters), with Fe(II) (H202, D204, and H259 shown in orange characters) and with GA4 (Q206, Y312, Y89, Y109, and R179 shown in green characters). c Overall structure of OsGA2ox3 tetramer composed of four subunits A–D (see details in the text). d Close-up view of the interface containing GA4 between subunit A and D. The GA4 in the interface and active site of subunit A and D are shown in blue and pink, respectively. Amino acids involved in interaction with the interface GA4 are highlighted, whereas the hydrogen bonds mediated by two water molecules (red spheres) are also indicated by dashed lines. e The gel filtration profile of WT-OsGA2ox3, WT-OsGA2ox6, and their mutation derivatives with or without GA4. 3Mu is OsGA2ox3 carrying C194A, K308A, and K313A. T, D, and M indicate the expected elution positions for tetramers, dimers, and monomers of OsGA2oxs. f Alignment of amino acids involved in multimerization of GA2oxs from rice and Arabidopsis, 194th, 308th, and 313th. The residue at the position of 308 is always shared by K or R. g BiFC analysis of multimer formation of WT-OsGA2ox3, WT-OsGA2ox6, and their mutation derivatives. YFP yellow fluorescent protein, Chl autofluorescence of chloroplasts. Scale bar = 10 μm.