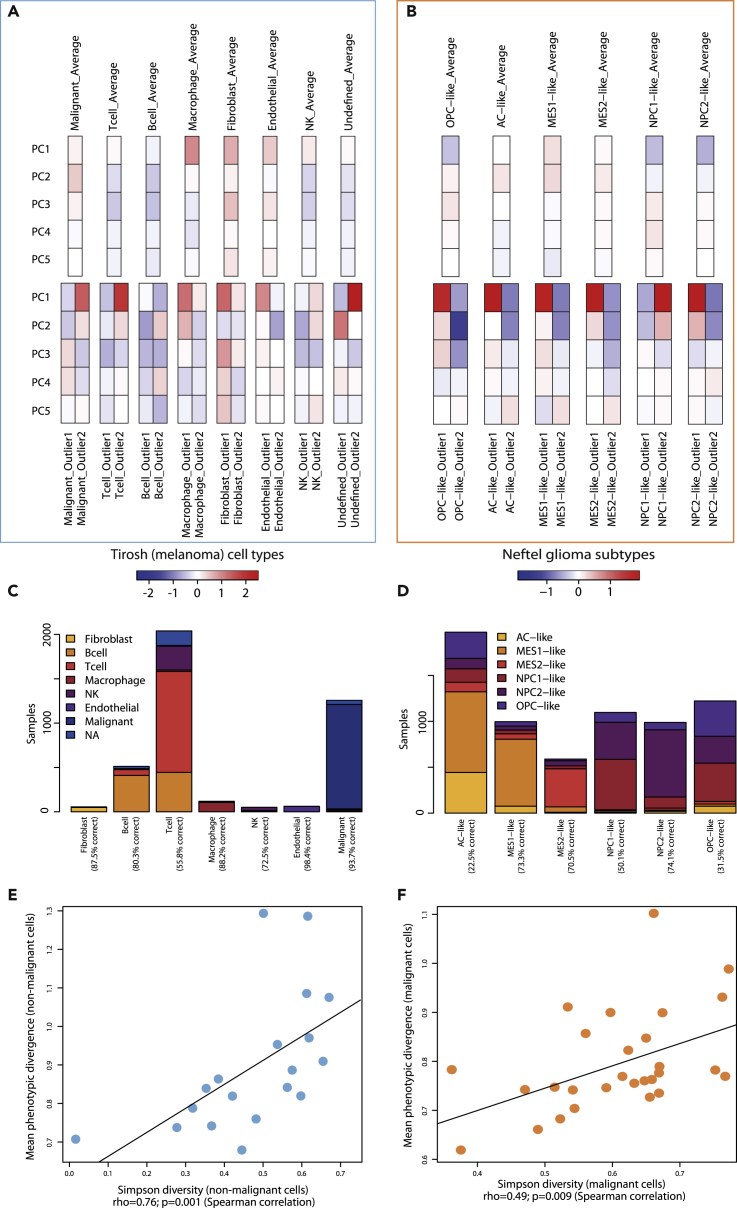
Figure 6.
Phenotypic Diversity in Populations of Known Structure
(A and B) PCA-based phenotypic profiles of (A) seven non-malignant cell types from the Tirosh et al. melanoma dataset and (B) six glioma subtypes from the Neftel et al. H3K27M-glioma dataset. Average profiles on top were obtained by averaging all cells from a given subtype across all patients. The outlier profiles at the bottom were obtained from the same-type cell pairs displaying the highest activity-based divergence for each cell type. Only the first five principal components are shown.
(C) Barplots showing the breakdown of how non-malignant cells from melanoma samples would be re-categorized, based on the average activity profiles of each category in the Tirosh melanoma dataset.
(D) Barplots showing the breakdown of how malignant glioblastoma cells would be re-categorized, based on the average activity profiles of each category in the Neftel dataset.
(E) Relationship between mean phenotypic divergence between non-malignant cells in the melanoma dataset and the Simpson diversity index calculated on the repartition of cells into the seven non-malignant classes.
(F) Relationship between mean phenotypic divergence between malignant cells in the glioma dataset and the Simpson diversity index calculated on the classification of cells into the six glioma subtypes. Black lines: linear models.