Figure 4.
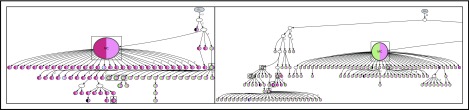
Lineage trees for patient 4 showing the N-gly site status of each subclone. The left panel represents the lineage tree of the first relapse event, whereas the right panel represents part of the lineage tree of the second relapse event. Each subclone is represented by a node; major clones are indicated in larger nodes. Nodes are split into 2 colors to represent the 2 N-gly sites found in the FR2 and CDR3 regions for patient 4 (Table 2). FR2 site is represented in the left half, and CDR3 site is represented in the right half, of the nodes. Nodes differing in color to the major clone represent subclones with different N-gly site codon sequences. Black represents absence of the FR2 or CDR3 site. Major clones between the 2 disease events have a different codon sequence in the FR2 N-gly site, indicated by the difference in color. Nodes in boxes represent shared subclones between events. White nodes represent subclones inferred to exist, but not detected through 454 sequencing. Germline nodes are in gray at the top of the tree, marked G.L. Numbers on branches indicate more than 1 mutation separating 1 node from the other. The full lineage tree of the second relapse event is provided in supplemental Figure 2A. MC, major clone.