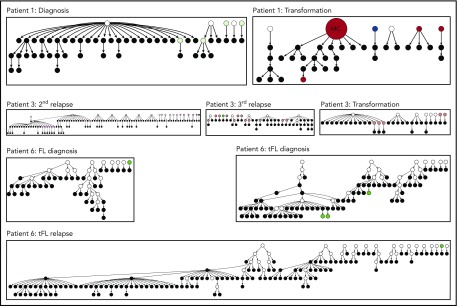
Figure 6.
Hierarchy of N-gly site-negative clones in relation to their direct ancestral clone for patients 1, 3, and 6. This diagram represents part of the lineage trees, as the majority of subclones (which are positive) are not shown and the interconnection between clones from a germline sequence is not depicted. Only the negative subclones, their parent clone, and their progenitor clones are depicted here to highlight their generation and contribution to the clonal repertoire within an event. For all patients, black nodes represent N-gly site-negative clones. White nodes represent clones not detected by the sequencing platform, but predicted to exist by the IgTree program. All white nodes are assumed to contain the N-gly site because of a number of other experimentally detected progeny clones being site-positive. Gray nodes in the patient 1 diagnosis event represent N-gly site-positive subclones that contain the same N-gly site-encoding codon sequence to the major clone. Dark red nodes in the patient 1 transformation represent N-gly site-positive subclones that contain the same N-gly site-encoding codon sequence to the major clone of the event. Pink nodes in patient 3 represent N-gly site-positive subclones that contain the same N-gly site-encoding codon sequence to the major clone of the event. Green nodes in patient 6 represent N-gly site-positive subclones that contain the same N-gly site-encoding codon sequence to the major clone of the event. Other colored nodes in trees represent N-gly site-positive clones, which contain a different codon sequence in the N-gly site compared with the major clone of the disease event. The full lineage trees can be requested from the corresponding author.