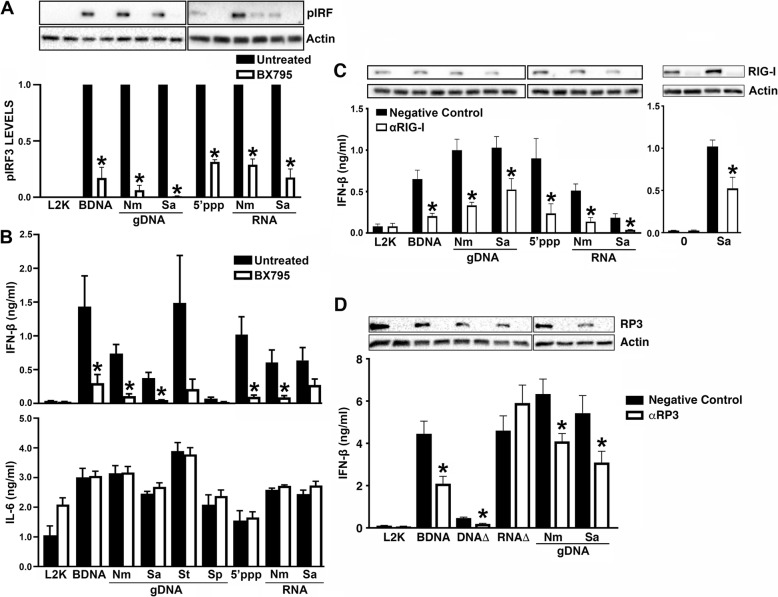
Fig. 4.
Bacterial components stimulate RIG-I-dependent signaling in hμglia human microglial cells. Cells were untreated or treated with 1 μM BX795, an inhibitor of TBK1/IKKε, for 3 h prior to transfection with 0.1 μg/ml BDNA, 1 μg/ml 5′pppRNA, 0.5 μg/ml bacterial genomic DNA (gDNA), or 1 μg/ml bacterial RNA (a, b). Total cell lysates were collected at 3 h and analyzed for protein expression of phosphorylated IRF3 (pIRF3) and β-actin via immunoblot analysis (a). Relative phosphorylated IRF3 expression was normalized to β-actin and is displayed graphically below the representative immunoblot. Cell supernatants were collected 24 h post transfection. IFN-β and IL-6 levels were quantified with specific capture ELISAs (b). Microglial cells were treated with scrambled siRNA or siRNA targeting RIG-I (αRIG-I) at a final concentration of 5 nM for 24 h. Cells were placed in fresh media for 24 h prior to transfection with 0.1 μg/ml BDNA, 1 μg/ml 5′pppRNA, 0.5 μg/ml bacterial gDNA, 1 μg/ml bacterial RNA, or infection with S. aureus at a MOI of 50 (c). Microglial cells were treated with scrambled siRNA or siRNA targeting RNA polymerase III (αRP3) at a final concentration of 5 nM for 24 h, then placed in fresh media for 24 h, and then transfected with 0.1 μg/ml BDNA, 0.5 μg/ml bacterial gDNA, or 5 nM DNA/RNA triangles (d). Cell supernatants were collected at 24 h post transfection and IFN-β levels were quantified by specific capture ELISA. Data are expressed as mean ± SEM for a minimum of three independent experiments. Asterisks indicate statistical significance compared to untreated condition as determined by Student’s t test or two-way ANOVA (p < 0.05)