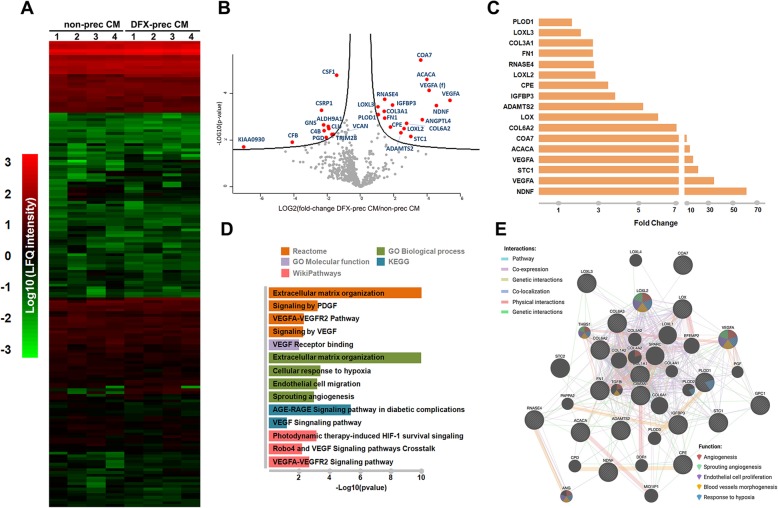
Fig. 9.
Mass spectrometry, functional enrichment, and network analysis of conditioned medium derived from non-preconditioned AD-MSCs and from DFX-preconditioned AD-MSCs. a Heatmap representation comparing the LFQ intensity (log10) of all 569 proteins recovered from conditioned medium derived from non-preconditioned AD-MSCs and from DFX-preconditioned AD-MSCs. b Volcano plot showing differentially expressed proteins between conditioned media derived from both conditions. The (f) associated to VEGF-α protein indicates a 111 amino acids (H0YBI8, UniProt) fragment different to the full length VEGF-α. c Histogram representing the set of 17 significantly overexpressed proteins in the conditioned medium derived from DFX-preconditioned AD-MSCs compared to the conditioned medium derived from non-preconditioned AD-MSCs. d Functional enrichment analysis (p value < 0.05) of the differentially expressed proteins according to Reactome, Gene Ontology (Molecular Function and Biological Process), KEGG, and Wiki Pathways databases. Enriched functional categories were selected based on their association with the angiogenesis process. e Network analysis of the selected overexpressed proteins according to different interaction categories and protein functions based on Genemania web server. The proteomic analysis was carried out using 4 different biological samples per condition