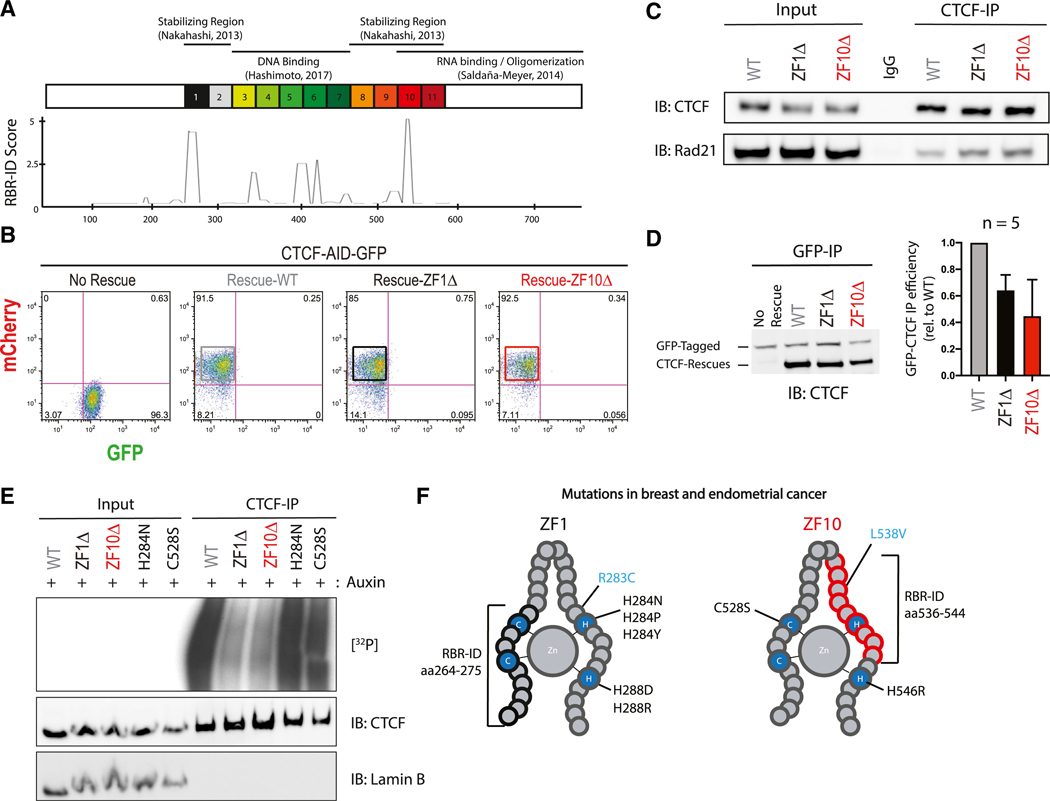
Figure 2. Deletions in ZF1 and ZF10 Independently Abolish CTCF Binding to RNA.
(A) Schematic representation of known domains of WT CTCF with its 11 zinc fingers being numbered (top); smoothed residue-level RBR-ID score (He et al., 2016), plotted along the primary sequence (bottom).
(B) FACS analysis highlighting percentage of GFP+ or mCherry+ CTCF-AID-GFP mESCs with or without rescue of CTCF: WT, ZF1Δ, or ZF10Δ.
(C) Immunoprecipitation of all rescue cell lines indicated and immunoblots for CTCF and Rad21.
(D) Representative image of GFP-CTCF incubated with each rescue, immunoprecipitated with a GFP antibody and blotted against CTCF (left); bar graph quantification of each rescue protein relative to the GFP-CTCF (n = 5) (right).
(E) PAR-CLIP of stably expressed WT and mutant CTCF in mESCs. Autoradiography for 32P-labeled RNA (top) and control western blot (middle and bottom).
(F) Schematic representation of ZF1 and ZF10 of CTCF; mutations found in breast and endometrial cancer that alter zinc binding are shown in black; mutations that do not alter zinc binding are in blue, and RBR-ID deletions are in brackets.