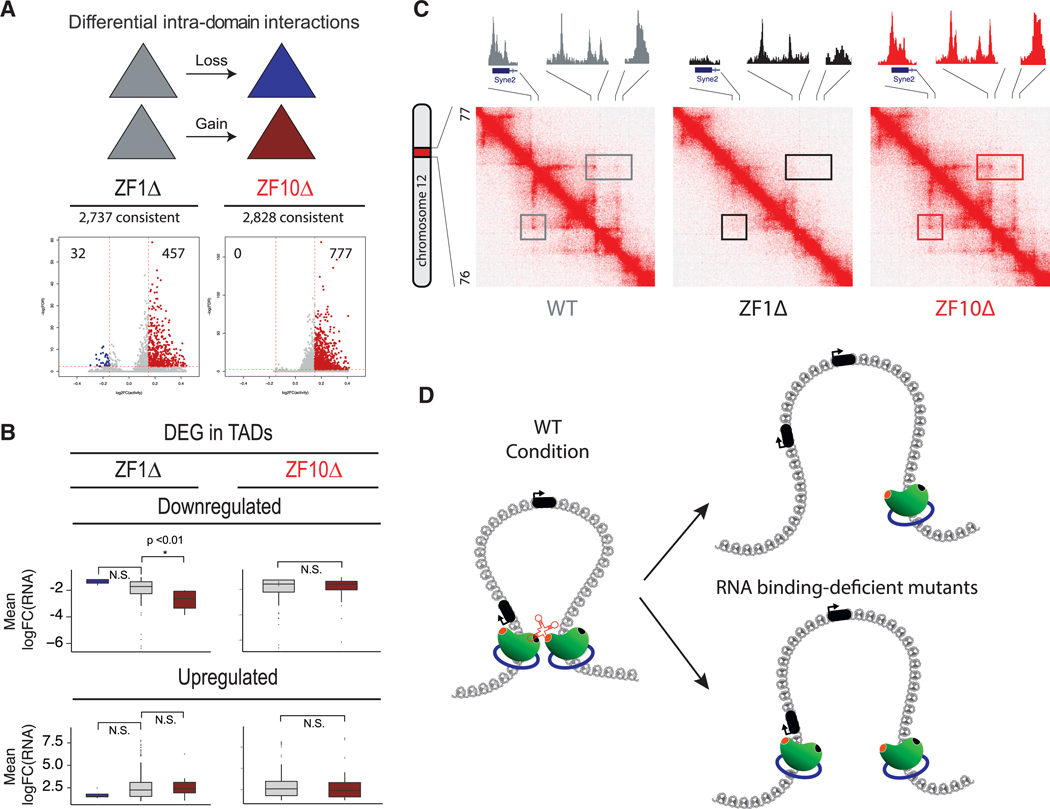
Figure 6. RBR Mutants Disturb Chromatin Loops.
(A) Intra-domain interaction changes in WT versus ZF1Δ and WT versus ZF10Δ for common domains. CTCF mutant rescues are associated with gain (red) and loss (blue) of intra-domain interactions.
(B) Boxplots representing the correlation between DEG and chromatin domains whose interactions are increased. Only downregulated genes for ZF1Δ are significantly correlated with increased intra-domain interactions, while all others are not significantly correlated.
(C) Representative contact matrix (at 5 kb resolution) show that the chromatin loop anchor on the left overlaps with the promoter of the Syne2 gene. In the WT rescue (left), the loop disappears in the ZF1Δ (middle) and is stable in ZF10Δ (right), while CTCF binding is lost only at the promoter and anchor for ZF1Δ. Syne2 is upregulated only under the ZF1Δ condition.
(D) Graphical representation of a chromatin loop formed by two CTCF proteins (green) and cohesin rings (blue), stabilized by an RNA in the WT condition (left diagram). Two outcomes are observed for the RNA binding-deficient mutants (ZF1Δ and ZF10Δ): (1) the loop is lost and a CTCF protein loses its binding to chromatin (top, right diagram), or (2) both CTCF proteins remain bound to chromatin yet the chromatin loop is still lost (bottom, right diagram).