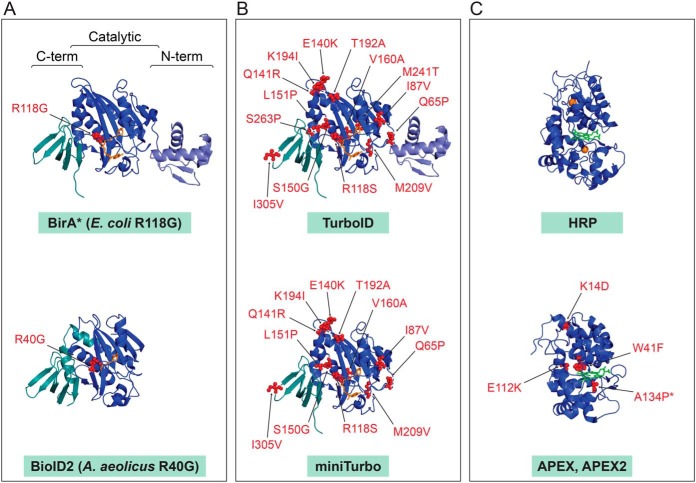
Fig. 4.
Structures of the main enzymes employed in PDB research. PDB files were downloaded from the RCSB Protein Data Bank and visualized in Pymol. Mutations over the wild type enzymes are indicated. A, Original enzymes used in BioID. Structures of the wild type E. coli BirA with biotinyl-5′-AMP (source PDB 2EWN) and A. aeolicus BPL with biotin (source PDB 3EFS) with position of the mutation decreases the affinity for biotinyl-5′-AMP intermediate. B, Molecularly evolved TurboID and miniTurbo displayed on the E. coli wild type BirA structure. C, Peroxidases. HRP with heme and calcium ions (source 1HCH) and APEX/APEX2 mutations displayed on the structure of soybean APX with heme (source PDB 1OAG) are indicated (note that APEX2 contains the additional A134P mutation compared with APEX).