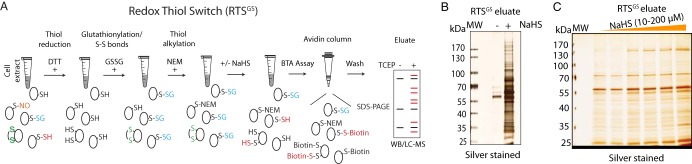
Fig. 6.
Identification of RTSGS targets by the BTA assay in vitro. A, Schematic of workflow to identify cysteine residues within the proteome, candidates for regulation by the RTSGS. Cell lysates were pretreated with TCEP to remove any reversible cysteine-based protein modifications, then incubated with GSSG to incorporate S-glutathionylation and disulfides to proteomes. The GSSG-treated protein extracts were treated with N-ethylmaleimide (NEM) to block any free thiol groups. After desalting, the lysates were exposed to H2S (NaHS) followed by the BTA assay. The RTS targets were analyzed in a SDS gel or by LC-MS. B, The BTA assay was performed to detect RTS targets in extracts from mouse liver. GSSG-treated mouse liver lysates were divided into two equal fractions, then treated without or with an H2S donor (NaHS, 2 mm) for 1h followed by the BTA assay. The eluates were analyzed by SDS-PAGE electrophoresis and silver staining of proteins. C, Mouse liver lysates (equal amount/treatment) were treated and analyzed as in B but with increasing concentrations of the H2S donor-NaHS (10, 20, 40, 80, 100, and 200 μm).