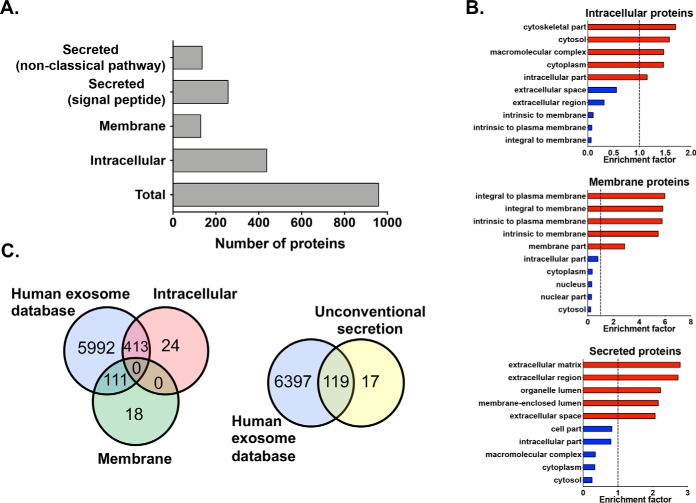
Fig. 2.
Classification of proteins by predicted topology, GOCC enrichment and exosome analyze. A, Number of proteins for each category (x axis). Using predictive software, all the proteins identified in mock- and hRSV-infected WD-PBEC cultures of each patient (n = 3) were combined and categorized into proteins secreted via a signal peptide or in a non-classical pathway, and cellular proteins with either intracellular or membrane localization. B, GOCC enrichment of the proteins predicted to be intracellular, membrane or secreted (classical and non-classical fashion), which identified the five most significantly enriched (>1, red bars) and depleted (<1, blue bars) cellular compartments in each protein group. x axis, gene ontology enrichment factor. Only GO classifications with a FDR < 1% are shown. C, Venn-diagrams showing the overlap between several categorises of secretome proteins and a human exosome database (Exocarta, http://www.exocarta.org/).